‘Cross’-Talking about Aspects in Cell Signalling Copyright © by Charlotte de Araujo is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, except where otherwise noted.
‘Cross’-Talking about Aspects in Cell Signalling Copyright © by Charlotte de Araujo is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License, except where otherwise noted.
1
This resource aims to provide an overview of concepts in cell signalling, introducing learners to the language of cellular pathways. The inquiry-solution blended style is presented to provide a mechanism to divide biochemical signalling pathways into more manageable modules, with the aim to keep the content more comprehensible.
1
Proteins undergo several post-translational modifications that extend their functions and alter their interactome. Phosphorylation is one of the most common and critical regulatory mechanisms for proteins. This unit aims to provide insights to the major enzyme groups involved in protein phosphorylation and dephosphorylation.
Upon successful completion of this chapter, you will be able to:
Kinases are a generic term for enzymes which are involved in transferring a phosphate group from one molecule (substrate) to another. In eukaryotes, conventional protein kinases introduce phosphate groups onto three key amino acids: serine (Ser), threonine (Thr) or tyrosine (Tyr).
i. Tyrosine kinases
ii. Serine/Threonine kinases
The name of the kinase reflects the specific amino acids which are phosphorylated. There are also dual-specific kinases which phosphorylate Ser, Thr as well as Tyr residues. As a guiding principle, serine and threonine protein phosphorylation are associated with large changes in protein conformation, whereas tyrosine protein phosphorylation is associated with altering the cellular localization of a protein. There are approximately 518 proteins identified as kinases within the human genome (Manning et al., 2002).
a. Hydroxyl (OH)
b. Thiol (SH)
c. Ether (-O-)
d. Alkene(C=C)
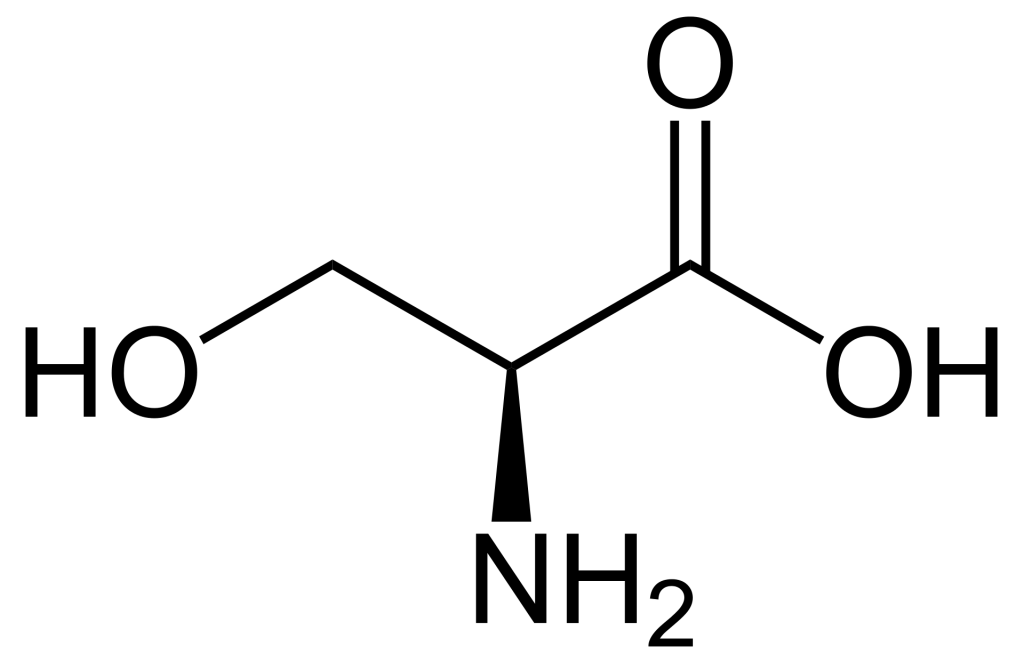
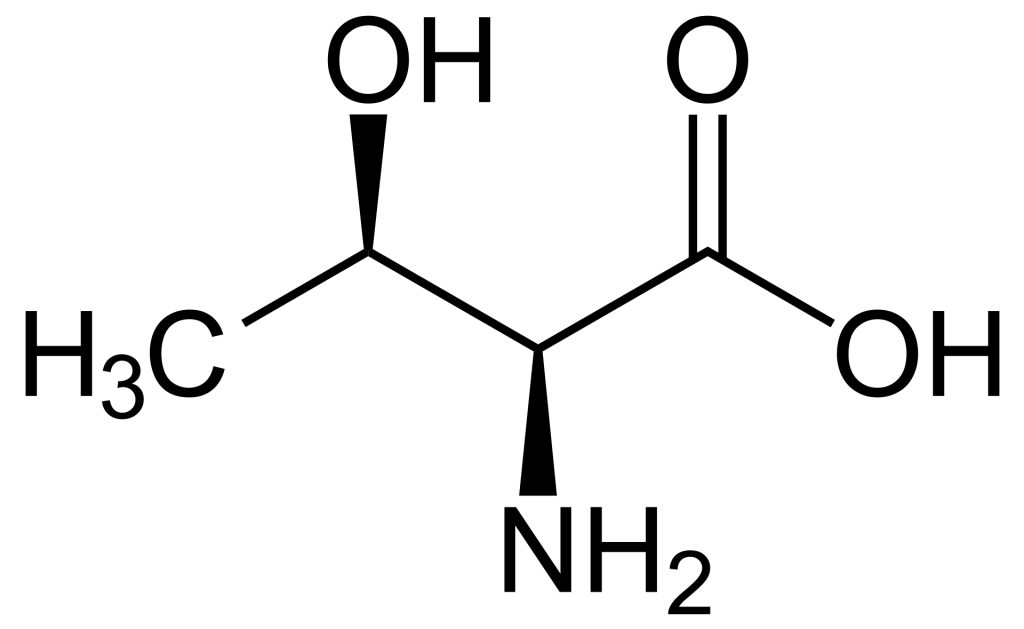
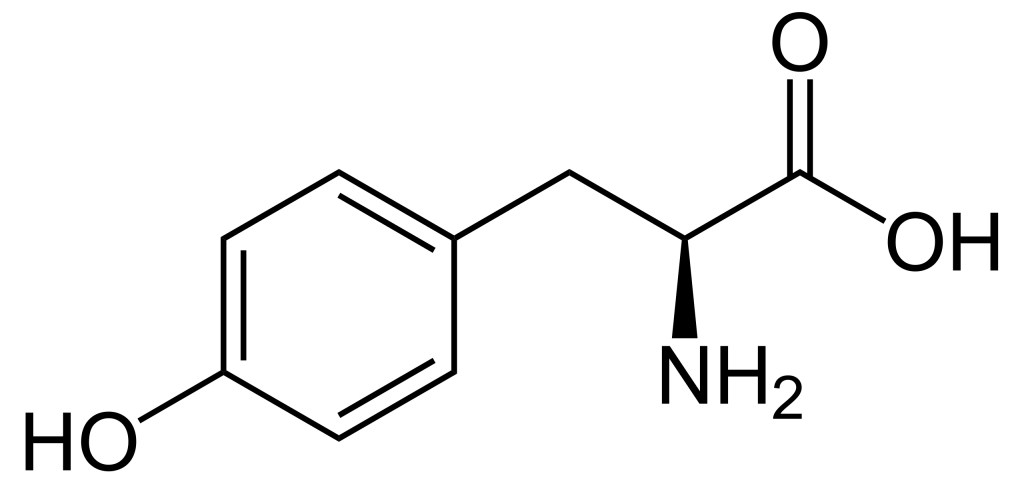
When comparing the structures of Ser, Thr, and Tyr, note that all of these amino acids possess a hydroxyl group as part of their side-chain group (Figure 1.1). A negatively charged phosphate group can be conjugated to these amino acids, generating a phospho-Ser, phospho-Thr, or phospho-Tyr respectively.
a. Increases protein catalytic activity
b. Decreases protein catalytic activity
c. Depends on the scenario
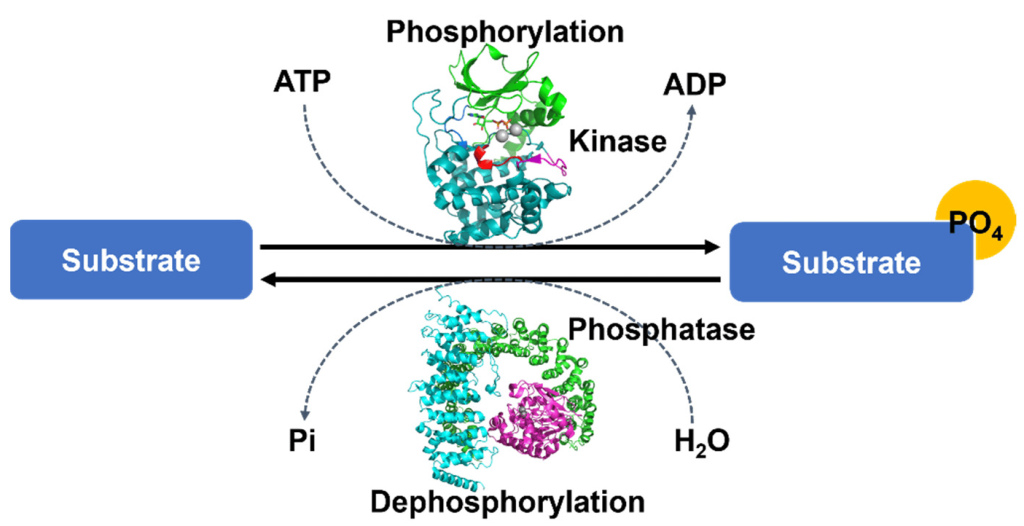
Phosphorylation of a protein (adding a phosphate group), may increase or decrease the catalytic activity of a protein depending on the consequent structural changes that impact the protein conformation (Figure 1.2). The phosphate group introduces a divalent negative charge at the protein site (distinct from any naturally occurring amino acid) which can drastically alter the physico-chemical properties of the protein.
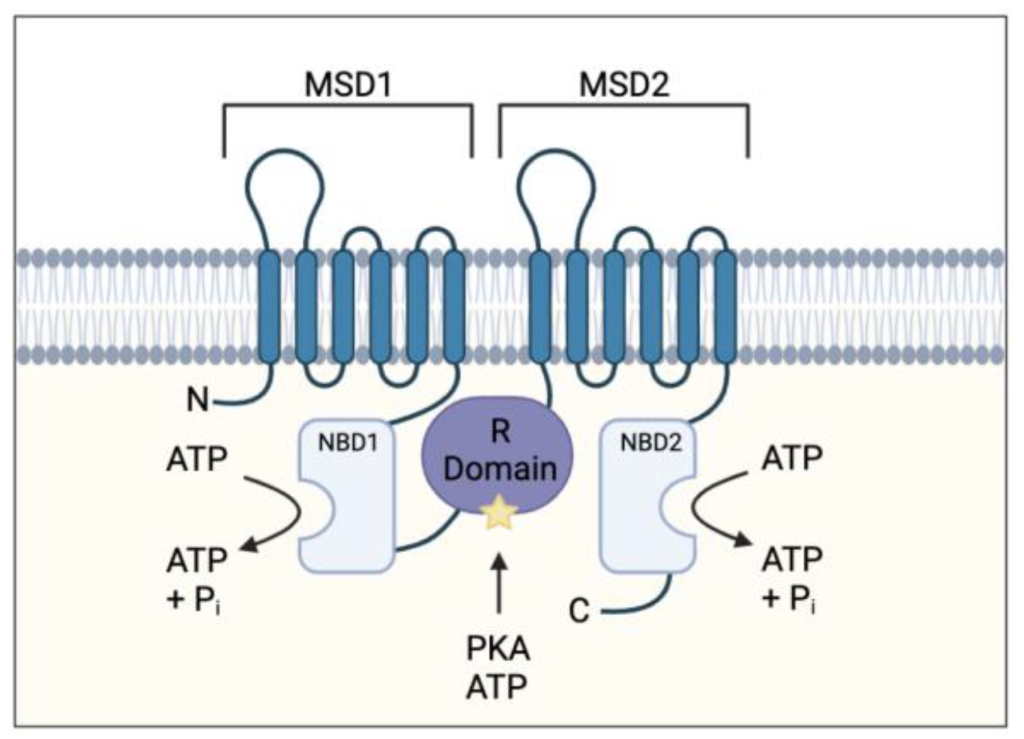
For example, the phosphorylation of a chloride ion channel protein called CFTR (cystic fibrosis transmembrane conductance regulator) occurs on a specific loop (called the R region) of the protein that normally blocks chloride ion flow through the channel (Figure 1.3). The resulting electrostatic perturbation as a result of phosphorylation of the loop enables it to engage with other regions of the protein, thereby removing it from sterically blocking pore access.
Furthermore, a subset of proteins are also prone to multi-site phosphorylation, where there are clusters of Ser or Thr available for phosphorylation. Increases in phosphorylation levels can result in a more pronounced effect. For example, the R region of CFTR contains at least ten phosphorylation sites, and progressive phosphorylation increases gating activity.
There are a number of ‘-omes’ categories in biology. The -ome suffix generally refers to a subset of biomolecules from a cell or organism. This can be referred to more broadly defined subsets, such as the genome (subset of genes within a cell) or proteome (subset of proteins within a cell). There are also more niche terms such as ‘kinome’ which is encompasses all of the kinases for an organism. The human kinome is often represented in a ‘kinome tree’ which represents a phylogenetic visual representation of the evolutionary relationships between the 518 different kinases (Manning et al., 2002).
Ser, Thr, and Tyr are common residues and are found in proteins. However, not all of these residues will be labelled with a phosphate group by the action of kinase. Kinases recognize a specific sequence containing a Ser/Thr/Tyr, called a ‘consensus sequence’. This peptide sequence provides a recognition motif for the kinase to bind and enables specific interactions with the kinase active site to position the appropriate Ser/Thr/Tyr into the pocket. For example, the recognition sequence of cAMP dependent protein kinase (PKA) is R-R/K-x-S-ϕ (where X is any amino acid and ϕ is a hydrophobic residue) (Kemp et al., 1977).
a. Enzymes which catalyze the hydrolysis of phosphoryl groups on protein substrates
b. Enzymes which catalyze the addition of phosphoryl groups on protein substrates.
c. Enzymes which catalyze the formation of disulfide bonds.
One of the advantages of phosphorylation as a protein post-translational event, is that the phosphate group may be removed in a reversible manner through dephosphorylation steps (Figure 1.2). This provides a mechanism of transient regulation, by effectively operating as a molecular switch to turn a protein ‘on’ or ‘off’. Protein phosphatases are enzymes which remove phosphate groups present on the amino acids Ser, Thr, or Tyr resulting in a return to the original protein side chain with a hydroxyl (-OH) group. An ortho-phosphate (Pi) group is subsequently released.
Analogous to kinases, there are two common classes of phosphatases which participate in cellular signalling pathways:
i. Ser/Thr phosphatases
ii. Tyr phosphatases
Tyrosine phosphatases are enzymes which catalyze the removal of phosphate groups from Tyr residues of protein substrates. Tyrosine phosphatases can be further classified into either receptor-like or non-transmembrane proteins. Receptor-like tyrosine phosphatases (RPTPs) will be discussed as part of the module on enzyme-coupled receptors. There are approximately 226 protein phosphatases (Liu & Chance, 2014).
An example of a member of a non-transmembrane protein phosphatase is Src homology-2 (SH2)-domain containing protein tyrosine phosphatase-2 or SHP-2, which is expressed in multiple tissues. SHP-2 is a specific example of a cytosolic phosphatase, composed of different protein domains including two tandem SH2 domains and a catalytic phosphatase domain (Qu, 2000). SHP-2 acts by dephosphorylating multiple proteins which generally has a negative role in blocking signalling pathways.
In the absence of a target protein, the phosphatase domain interacts with the N-terminal SH2 domain of SHP-2. This interaction maintains the protein in a closed conformation and prevents access of substrates to the phosphatase site. Therefore, SHP-2 is unable to carry out phosphate hydrolysis reactions.
In the presence of a target (e.g. a protein with phosphorylated tyrosine), the phosphorylated target will bind SH2 domain of SHP2. This binding event triggers a conformational change, altering interactions between SH2 domain and catalytic domain. The active site becomes more accessible, enabling protein binding at the catalytic phosphatase domain and eventual removal of the phosphate group from the target protein. The products are released to reset the catalytic cycle.
Within the tyrosine phosphatase group of proteins, there is another sub-family, referred to as DUSPs (dual-specificity phosphatases) which are phosphatases hydrolyzing phosphate groups primarily from Tyr residues. However, they also possess Ser/Thr phosphatase activity.
Harwood, K. H., McQuade, R. M., Jarnicki, A., & Schneider-Futschik, E. K. (2021). Anti-Inflammatory Influences of Cystic Fibrosis Transmembrane Conductance Regulator Drugs on Lung Inflammation in Cystic Fibrosis. International Journal of Molecular Sciences, 22(14). https://doi.org/10.3390/ijms22147606
Kemp, B. E., Graves, D. J., Benjamini, E., & Krebs, E. G. (1977). Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. The Journal of Biological Chemistry, 252(14), 4888–4894. https://www.jbc.org/article/S0021-9258(17)40137-2/pdf
Liu, Y., & Chance, M. R. (2014). Integrating phosphoproteomics in systems biology. Computational and Structural Biotechnology Journal, 10(17), 90–97. https://doi.org/10.1016%2Fj.csbj.2014.07.003
Manning, G., Whyte, D. B., Martinez, R., Hunter, T., & Sudarsanam, S. (2002). The Protein Kinase Complement of the Human Genome. Science, 298(5600), 1912–1934. https://doi.org/10.1126/science.1075762
Qu, C. K. (2000). The SHP—2 tyrosine phosphatase:Signaling mechanisms and biological functions. Cell Research, 10(4), 279–288. https://doi.org/10.1038/sj.cr.7290055
Seok, S.-H. (2021). Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life, 11(9). https://doi.org/10.3390/life11090957
2
The fundamentals of biochemical processes in the body rely on intricate and highly complex enzymatic signalling cascades. These signalling pathways may employ receptors situated at the cell surface (e.g. enzyme-coupled receptors, G-protein coupled receptors) or nuclear receptors. This unit will examine the structural and functional properties of various enzyme-coupled receptors.
The term enzyme-coupled receptors can be examined as bringing together the two discrete components – receptors and enzymes. These proteins are receptors that also possess enzyme activity – the ability to catalyze a reaction. Receptors are transmembrane proteins, and they interact with a signal or ligand. This interaction occurs on the extracellular side of the cell and leads to a conformational change in the protein leading to enzymatic activity on the intracellular side of the cell. In this way, enzyme coupled receptors enable cellular signalling processes.
There are six major classes of enzyme-coupled receptors (Alberts, 2002).
I. Receptor tyrosine kinases (RTK)
II. Tyrosine-kinase-associated receptors
III. Receptor serine/threonine kinases
IV. Histidine-kinase-associated receptors
V. Receptor guanylyl cyclases
VI. Receptor-like tyrosine phosphatases
Upon successful completion of this chapter, you will be able to:
There are 58 RTKs known to exist within the human genome and they respond to different extracellular signals (Robinson et al., 2000). However, all RTKs possess common structural domains: an extracellular ligand binding domain, a transmembrane helix, a juxtamembrane region, and a tyrosine kinase (TK) domain.
RTKs are integral proteins that extend through the plasma membrane (via the transmembrane domain which is a single helix). The N-terminal region of the receptor is located in the extracellular region, while the C-terminal portion is situated in the cytoplasm.
The N-terminal region of the RTK contains the extracellular ligand binding site, which is the location of where an external signal will engage with the protein. This extracellular region is highly variable between different RTKs, and may include Cys-rich regions, Leu-rich segments, or immunoglobulin-like motifs. This helps maintain diversity for recognition of different external biochemical signals (Lemmon & Schlessinger, 2010).
The C-terminal region exists within the interior of the cell (the cytosol) and includes the conserved tyrosine kinase domain and juxtamembrane region. This kinase domain carries out the phosphorylation activity of the RTK whereas the juxtamembrane is highly flexible and important for regulatory roles, often leading to auto-inhibition via contacts with kinase domain.
a. Ser
b. Thr
c. Tyr
d. Option a and b
RTKs phosphorylate Tyr residues on protein substrates.
a. α
b. β
c. γ
d. α and β
e. β and γ
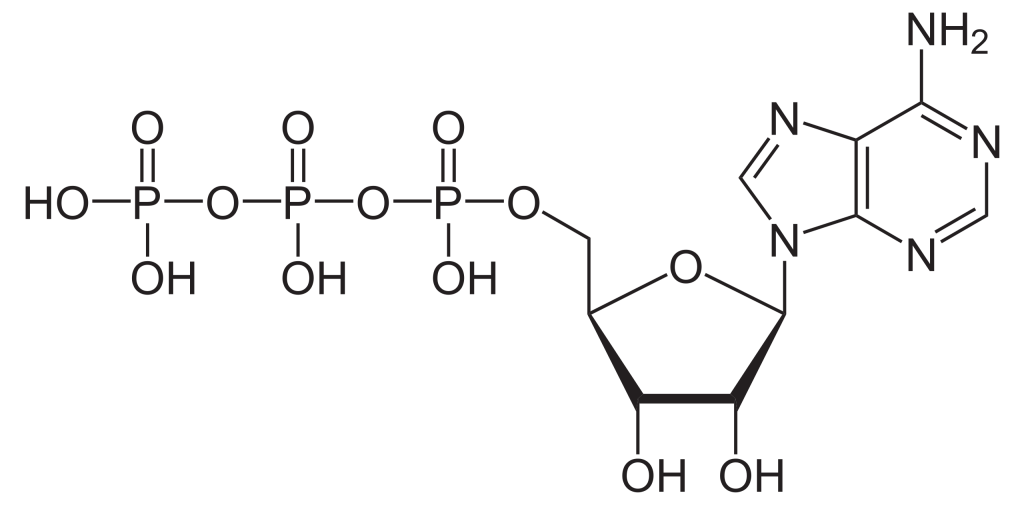
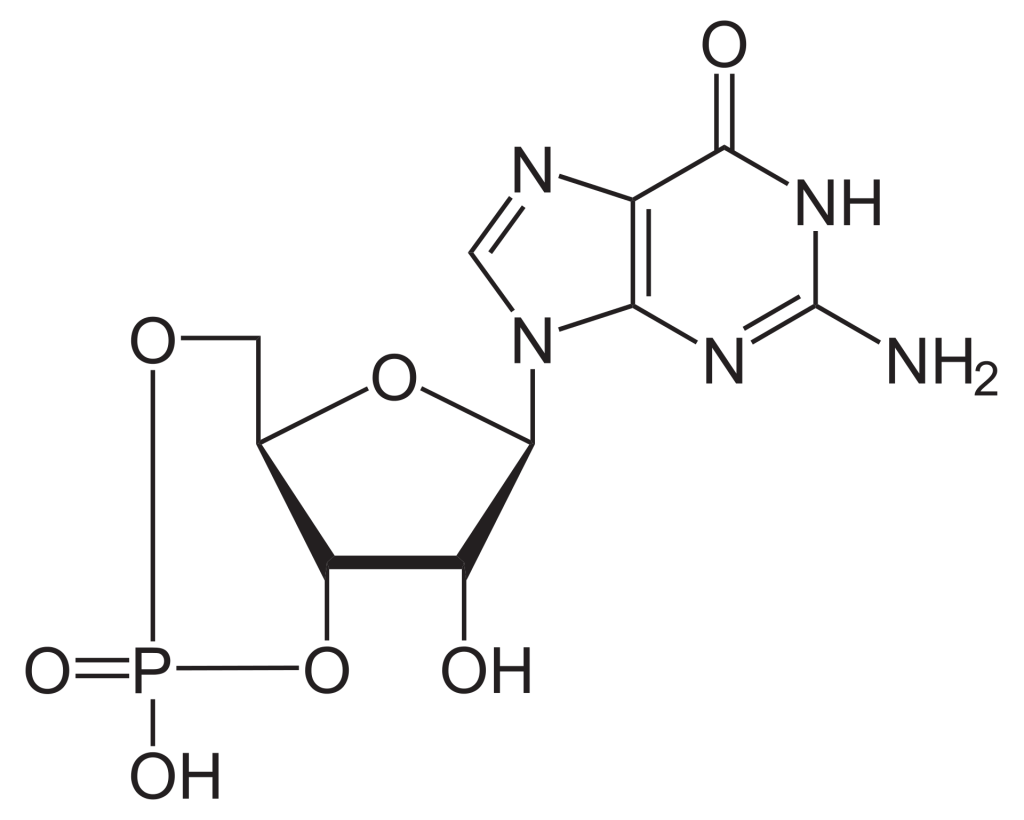
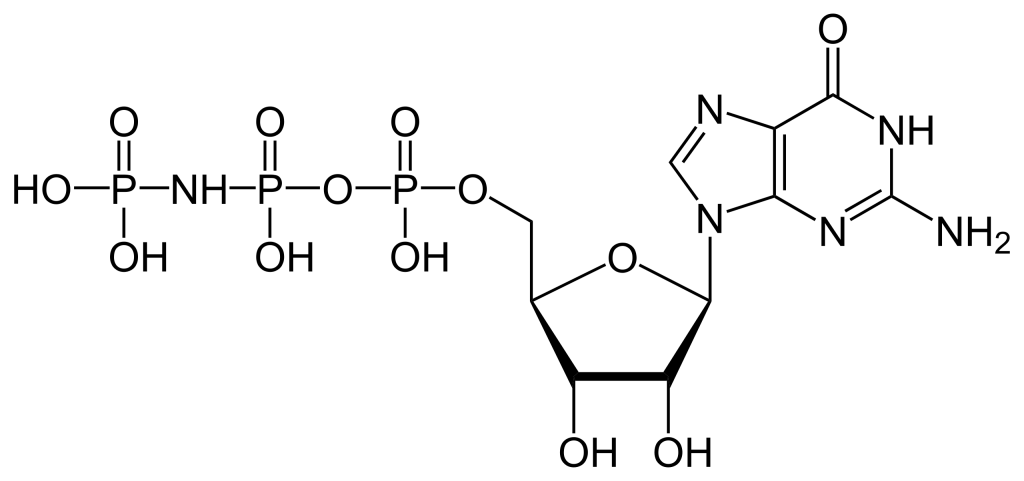
To build the nucleotide, ATP, three essential components are required: a five carbon ribose sugar, a nitrogenous base (adenine) and three phosphate groups which are labelled, α, β and γ (Figure 2.1). The α phosphate is positioned closest to the sugar group while the γ phosphate is furthest away. RTKs transfer a phosphate group from ATP adding a phosphate group to Tyr on protein substrates. The γ phosphate group is transferred during the phosphorylation step to a protein substrate. This is a highly exergonic reaction.
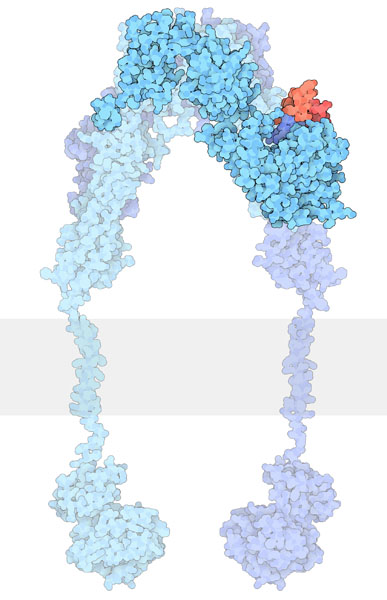
In the ‘inactive state’ RTKs generally exist as a single protein which is referred to as a monomer. However, the ‘active’ form requires two RTK proteins to come in close proximity which is called a dimer (Figure 2.2). However, there are exceptions. For example, the insulin receptor (INSR or IR) also exists as a dimer in the inactive form (Figure 2.3) due to a cysteine disulfide bridge linkage between the monomeric species.
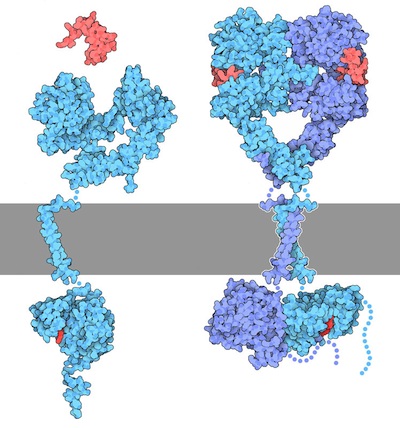
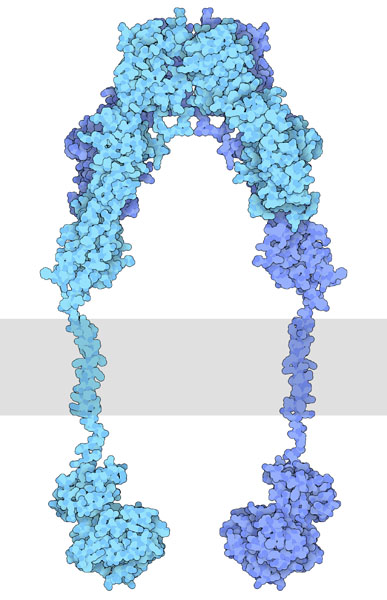
For this mechanism, we will consider RTKs that are initially in an inactive monomeric form. When an appropriate ligand binds to the receptor at the extracellular ligand binding domain, this results in a conformational change which converts the inactive RTK to the active form and facilitates bringing two monomer RTKs together. These two monomers are positioned in close proximity to each other, leading to the formation of a dimer.
Recall, the TK domain is located in the cytosolic portion of the receptor. Once the dimer is formed, the TK domains are positioned to carry out ‘trans-autophosphorylation.’ This is where a TK domain on one monomer phosphorylates selective Tyr residues located on the opposite monomer. This “trans-phosphorylation” converts the kinase domain into an active kinase domain, and the inactive RTK is converted to an active RTK.
Yes, ligand binding (e.g. epidermal growth factor, EGF) to the ligand binding domain of the receptor (e.g. epidermal growth factor receptor – EGF receptor) promotes conformational changes, eventually activating the RTK enzymatic activity (Figure 2.2). In this way, signals external to the cell are transmitted to the cellular interior, without the molecule/hormone crossing the membrane.
In this mutated RTK, the transmembrane and cytoplasmic domain are assumed to be functionally and structurally intact. However, as the ligand binding domain absent, there is no regulatory response and the RTK is no longer sensitive to its cognate ligand. In this case, the receptor will be in a perpetual ‘on’ state or constitutively active state. A negative consequence is a receptor which is ligand-independent can lead to hyper-phosphorylation and hyper-activation of downstream RTK pathways. In fact, 30% of human cancers have been demonstrated to have hyperactivated or overexpressed RTKs (Ségaliny et al., 2015).
a. Inhibit EGFR activity.
b. Activate EGFR activity.
As previously discussed, SHP2 is a phosphatase. In close proximity to EGFR, it would likely remove the phosphate group from EGFR (Lemmon & Schlessinger, 2010) which would reduce the activity of this RTK.
Following trans-autophosphorylation, RTKs possess phospho-Tyr residues. These residues function as sites to enable the binding and recruitment of specific signalling proteins. The regions on the RTK which contain the phospho-Tyr residues are also referred to as docking sites or binding sites. Signalling proteins which bind to this region possess phospho-Tyr-binding domains.
a. Extracellular region
b. Transmembrane portion of the receptor
c. Intracellular region
Some RTKs such as INSR exist as dimers in the inactive form. Similar to other RTKs, such as EGFR, the ligand binding domain is located in the extracellular domain (Figure 2.4).
a. Ser
b. Met
c. Cys
d. Cys and Met
e. Cys, Met, Ser
INSR contains disulfide bonds which are formed from two Cys residues.
Similar to other RTKs, the ligand binds to the receptor and this interaction results in a conformational change which activates the tyrosine kinase domain. Each monomer (in blue) can phosphorylate the opposing monomeric unit (Figure 2.4). The active tyrosine domain carries out the ‘autophosphorylation’ step, phosphorylating the other unit at tyrosine residues.
a. INSR activity would be enhanced.
b. INSR activity would decrease.
c. No effect on INSR activity.
DTT is a reducing agent, which would target the disulfide bonds, catalyzing their reduction. Treatment with DTT would be predicted to decrease receptor activity (Pike et al., 1986).
a. Cys
b. Met
c. Tyr
INSR is a RTK, where Tyr residues are phosphorylated.
INSR (in the active state) phosphorylates other cellular targets, including a common target Insulin Receptor Substrate (IRS).
Proteins associated with RTK pathways often possess SH2 (Src homology region) domains or PTB (phosphotyrosine-binding) domains for phosphotyrosine binding. To illustrate an example, IRS-1 binds the phosphorylated receptor using its PTB domain. Another case involves Grb2 which binds phophotyrosine containing motifs through an SH2 domain.
The SH2 domain is a protein domain that serves to bind phospho-tyrosine residues. The SH2 domain has two sub-pockets which are formed by two alpha helices that are separated by a central β sheet (comprised of three β strands). The phospho-tyrosine residue inserts into one sub-pocket (called the pY pocket) on one side the central β sheet, and residues C-terminal to the phospho-tyrosine insert into the other sub-pocket (called the pY + 3 pocket) on the other side of the β sheet (Bajusz et al., 2023). These flanking residues confer specificity via key amino acid side chains. Therefore, different SH2 domains will recognize phospho-tyrosine containing proteins within the context of the surrounding amino acids. This mechanism of action of a SH2 domain is often compared to a plug and a two-pronged socket (Waksman et al., 1993).
Insulin (ligand) binds to the cognate receptor, INSR, located at the cell surface of specific tissues (Figure 2.5). This results in a conformational change and subsequent auto-phosphorylation of specific Tyr residues on INSR. The activated INSR phosphorylates targets including the adaptor protein insulin receptor substrates (e.g. IRS-1). IRS-1 (in its phosphorylated state) serves as a docking site for a kinase, Phosphatidylinositide 3-kinase (PI3K). The enzyme PI3K catalyzes the phosphorylation of Phosphatidylinositol 4,5-bisphosphate (PIP2) to Phosphatidylinositol-3,4,5-triphosphate (PIP3). PIP3 activates another kinase, PDK1 (PIP3-dependent protein kinase). In the active form, PDK1, phosphorylates other kinases including Protein kinase B (PKB, also known as Akt1). Similarly, mTORC2 also phosphorylates and activates PKB. Continuing the series of phosphorylation events, PKB, phosphorylates targets including glycogen synthase kinase 3 (GSK3). In this case, phosphorylation inactivates the enzyme GSK3. In the phosphorylated form, GSK3, is unable to inactivate glycogen synthase (GS). GS is an enzyme which participates in converting glucose into glycogen.
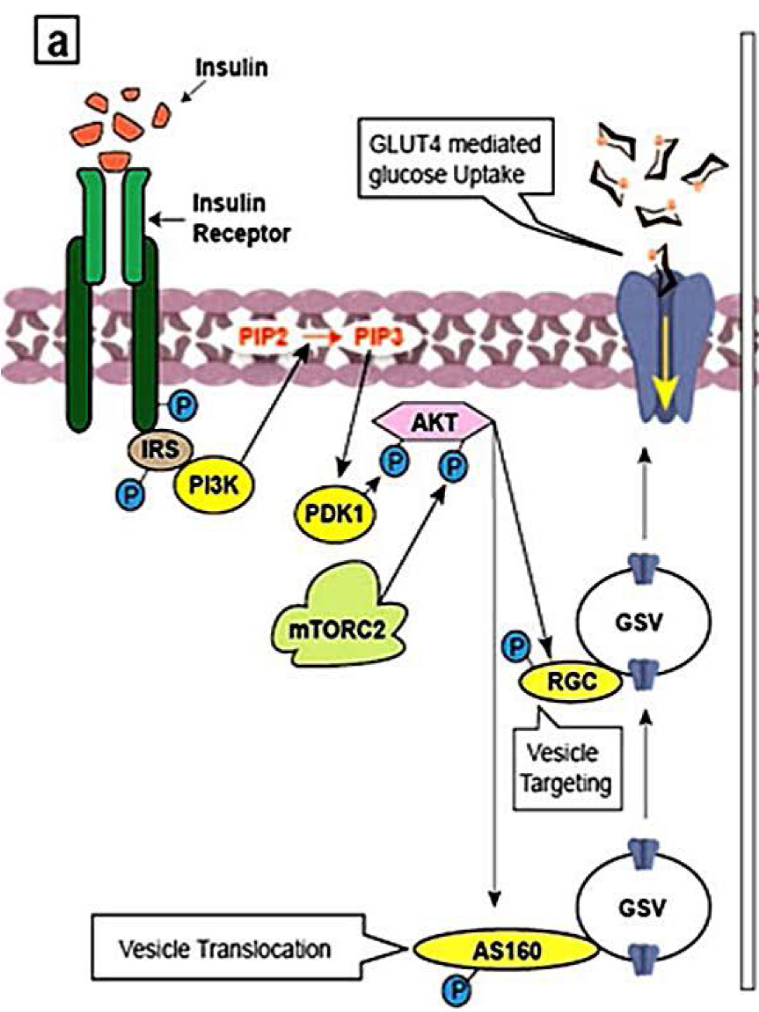
Here the term mTORC2 will be parsed further. TOR is a protein kinase and the mammalian form is referred to as mTOR or mammalian target of rapamycin. Together two different protein complexes mTORC1 and mTORC2 form mTOR. As a Ser/Thr protein kinase, mTOR phosphorylates Ser or Thr (whereas the original phosphorylation events were phospho-Tyr driven). The protein complex, mTORC2 is classically activated by variety of elements ranging from amino acids and growth factors such as insulin (Tato et al., 2011; Yoon, 2017) (Figure 2.5).
PKB also recruits and activates insulin-sensitive GLUT4-containing vesicles to promote glucose transport. PKB phosphorylates the GTPases, RAB GAP AS160 and RAL-GAP complex (RGC) (Figure 2.5) (Sayem et al., 2018). These are GTPases that promote the transport of GLUT4 vesicles to the plasma membrane. GLUT4 vesicles contain glucose transporter (GLUT4) which are incorporated in the plasma membrane. These proteins facilitate the transport of glucose from the blood into the cell and are known for making cells “permeable” to glucose.
Ras refers to a family of proteins which are small monomeric GTPases. Ras consists of isoforms (e.g. H-ras, K-ras, N-ras). The GTP binding protein Ras was identified in Rat sarcoma, hence forming the basis for the name.
The light switch (Figure 2.6) toggles between the ‘ON’ and ‘OFF’ options, where light appears in the ‘ON’ state and light is turned off in the ‘OFF’ state.
Similarly, when Ras is bound to the nucleotide guanosine triphosphate (GTP) it is in the active state.
Ras + GTP → active or ‘ON’
Whereas, when Ras is bound to the nucleotide guanosine diphosphate (GDP), Ras is in the inactive state.
Ras + GDP → inactive or ‘OFF’
Therefore, Ras is analogous to a switch, existing in two forms (active and inactive) (Figure 2.6).

GTP is a nucleotide composed of three key components: five carbon sugar (ribose), nitrogenous base (guanine) and three phosphate groups. GDP is a nucleotide formed from the same components except that it possesses two phosphate groups.
Guanosine nucleotide-exchange factors (GEFs) are typically proteins which catalyze the exchange of GDP to GTP. When Ras is bound to GDP, GEF catalyzes the switch of GDP to GTP.
GTPase-activating proteins (GAPs) support the hydrolysis of GTP to GDP. When Ras is bound to GTP, GAPs catalyze the switch from GTP to GDP. Ras is now bound to GDP.
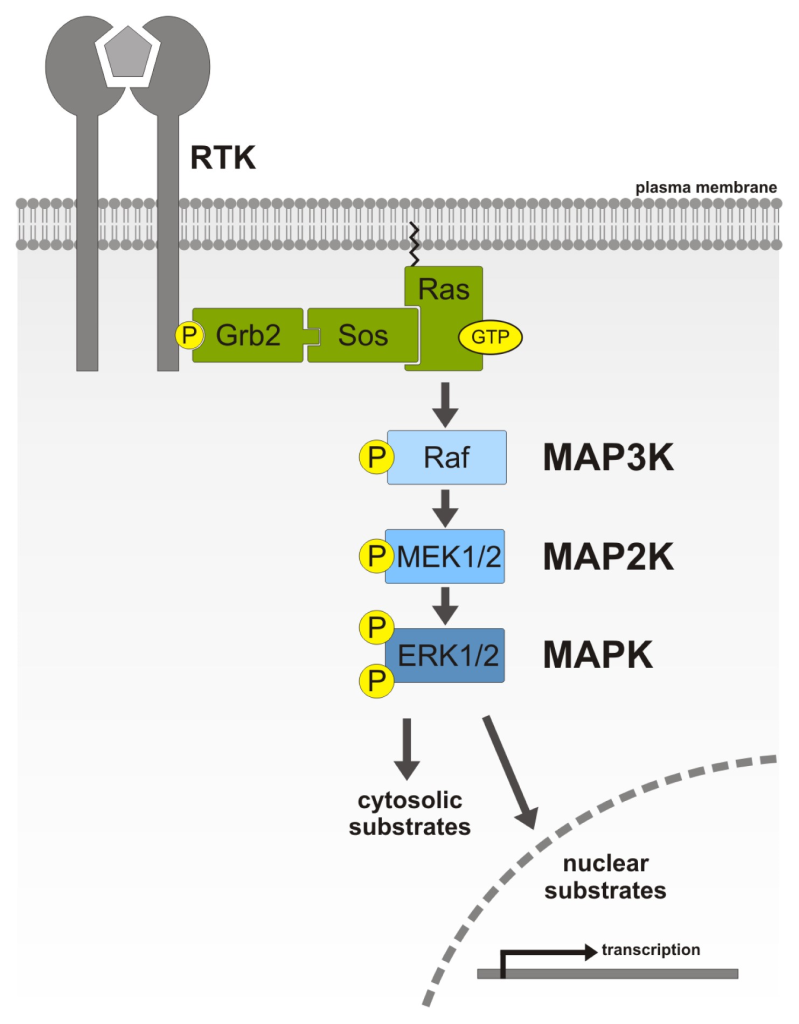
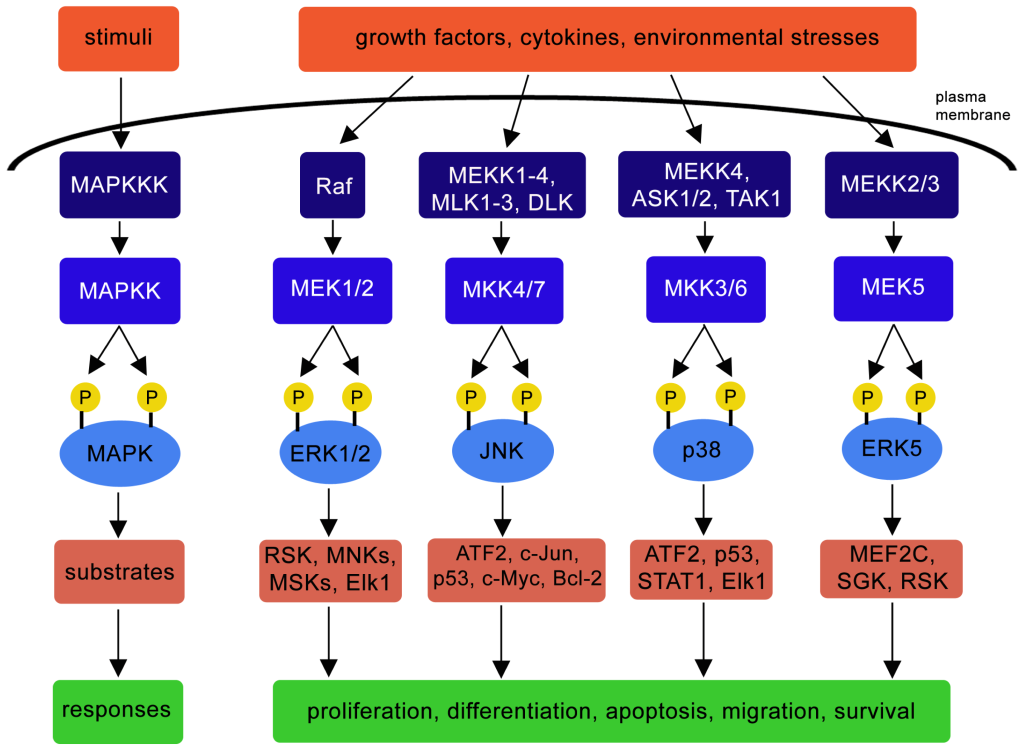
Mitogen-activated protein kinases (MAPKs) or MAP Kinases are an ubiquitous group of enzymes present in eukaryotes (Figure 2.7). In mammals the following representative groupings (i-iii) are members of the MAPK family (Morrison, 2012).
i. c-Jun NH2-terminal kinases (JNKs)
ii. Extracellular signal-regulated kinases (ERKs)
iii. p38 mitogen-activated protein kinases (p38s)
a. Ser
b. Thr
c. Tyr
d. Option a or b
e. All of the above are true
Members of the MAPK family phosphorylate Ser or Thr residues.
Members of the MAPKK and MAPKKK family phosphorylate both Ser or Thr.
A representative MAPKKK, c-Raf (Figure 2.7) phosphorylates specific Ser residues on MEK. Members of the MAPKK family phosphorylate Ser, Thr residues of the MAPK family.
Within the literature, there are various names used to describe the following kinases. Match the following kinase with alternative names used to describe them (Table 1).
| Kinase | Alternative Name |
| MAPK | MAP kinase |
| MAPKK | MAP2K or MAP kinase kinase |
| MAPKKK | MKKK or M3K or MAP3K or MAP kinase kinase kinase |
| MAPKKKK | MAP4K or MAP kinase kinase kinase kinase |
Within the MAPK signalling pathway, there are a series of phosphorylation events where MAPKKK initially phosphorylates MAPKK, which phosphorylates MAPK (Figure 2.7). A relay or ‘cascade’ of phosphorylation reactions takes place (Meister et al., 2013). MAPK will then proceed to phosphorylate additional proteins such as transcription factors.
MEK phosphorylates both Ser/Thr or Tyr residues on the protein substrate (Table 2). For example, MEK phosphorylates specific Thr and Tyr residues on Erk (Zheng & Guan, 1993).
| Component | Role |
| Ras | Monomeric G protein |
| Raf | Ser/Thr Kinase |
| MEK | Dual-specificity protein kinase |
| Erk | Ser/Thr Kinase |
MAPK pathway is involved in a spectrum of biological outcomes including promoting cell proliferation, cell differentiation, apoptosis, migration, cell survival depending on the kinase and corresponding substrates (Figure 2.8) (Osaki & Gama, 2013).
For example, a signal (e.g. cytokine) may activate the MAPKKK, apoptosis signal-regulating kinase 1 (ASK1) (Ichijo et al., 1997) (Figure 2.8). A series of phosphorylation events ensue, such as the phosphorylation of MKK3 and MKK6 (both MAPKKs). Both phosphorylated MAPKKs can activate p38 (a MAPK) through phosphorylation events. Activated p38 targets a number of substrates such as activating transcription factor 2 (ATF2), p53, STAT1, Elk1 resulting in different cellular responses (Figure 2.8). Targeted Ser phosphorylation by p38 of the tumor suppressor p53 promotes one cellular response – apoptosis (Bulavin et al., 1999).
IRS in the phosphorylated state recruits Grb2 (Growth factor receptor-bound protein 2). Grb2 binds to IRS-1 via the Grb2 SH2 domain. Following this, Grb2 recruits the protein Sos. Sos is a GEF, and promotes the switch between the GDP bound and GTP bound state in G-proteins. One of the key proteins that Sos associates with is Ras, and Ras is converted to the GTP bound state.
Recall, Ras-GTP is now in the active state and Ras can activate MAPKKK (Raf-1) (Figure 2.7). This leads to a series of phosphorylation steps part of the MAPK cascade.
This interaction is mediated by an ‘anchor’ composed of:
a. Polypeptide chain
b. Hydrophobic lipid group (e.g. farnesyl group)
c. Ubiquitin tag
d. Hydrophilic sugar group
As part of the MAPK pathway, Ras is linked to the plasma membrane and is situated on the cytosolic face via a lipid anchor called a farnesyl group. The anchor is shown by the red structure embedded as part of the plasma membrane (gray) (Figure 2.9) located at the C-terminus (Zhou et al., 2018).
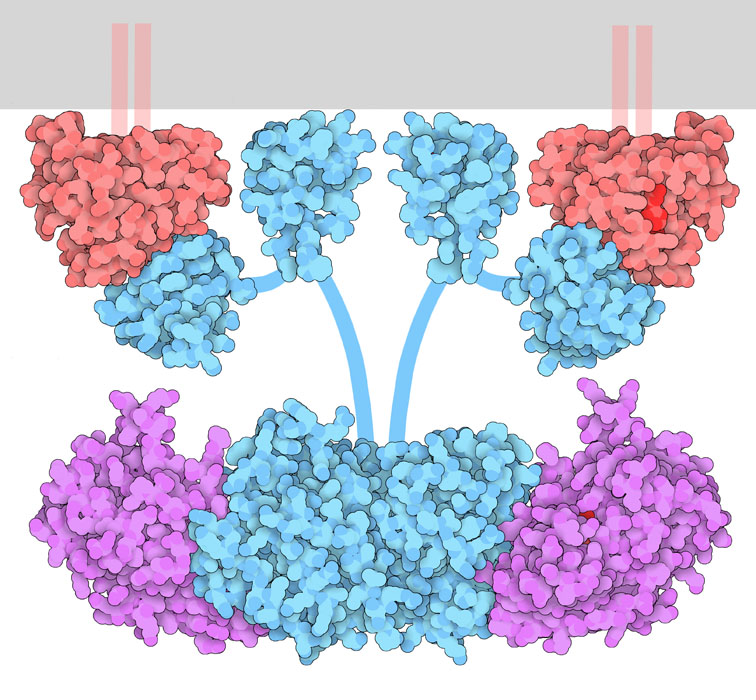
a. Sos
b. Ras
c. INSR
d. Grb2
Sos is an example of a guanine nucleotide exchange factor (GEF).
In signalling pathways, adaptor proteins help to mediate interactions between other proteins. While adaptor proteins contain different protein domains (e.g. SH2 domains), catalytic activity is usually not ascribed to adaptor proteins. Grb2 is an example of an adaptor protein which participates in multiple signalling pathways, including the insulin signalling pathway. Grb2 contains an SH2 domain and two SH3 domains. The SH2 domain serves to bind an activated phosphotyrosine molecule, and SH3 domains recognize proline-rich sequences. As an adaptor protein, Grb2 serves as a link between the Sos protein (via SH3 domains) and the RTK (via the SH2 domain).
a. Halting the MAPK pathway
b. Activation of the MAPK activity
c. No effect of the pathway
Sos is a cytosolic protein. Grb2 binds to the proline-rich domains of Sos through the two SH3 domains (Buday et al., 1994). Following these interactions, Sos localizes towards the plasma membrane and is positioned near (the lipid-bound) Ras thereby promoting nucleotide exchange. However, in some situations, phosphorylation of Sos will disrupt the Sos-Grb2 complex, causing dissociation of these proteins, ultimately negatively regulating the MAPK pathway (S. Corbalan-Garcia S.-S. Yang & Bar-Sagi, 1996).
To highlight the role of scaffold proteins, consider the role of scaffolding when constructing large buildings. The scaffolding material provides support during the assembly process. In an analogous manner, KSR is a scaffold protein, enabling a surface for various kinases to assemble (e.g. Raf, MEK, MAPK) and position in close proximity to each other (Morrison, 2001).
Both classes of receptors are capable of carrying out tyrosine phosphorylation and have intracellular, extracellular, and transmembrane domains. However, RTKs directly harbour kinase domains in the cytosolic regions, whereas tyrosine-kinase associated receptors will associate with an independent kinase protein. The enzymatic catalytic activity is separate from the C-terminal domain of the receptor in tyrosine-kinase associated receptors.
a. Covalently
b. Non-covalently
The tyrosine-kinase domains are not directly part of the primary sequence of the receptor, and are separate entities. For instance, cytokine receptors do not have endogenous tyrosine kinase domains. Rather the tyrosine kinase activity is associated noncovalently with Janus kinases (JAKs).
Cytokines are the signalling molecules activating corresponding cytokine receptors. They are proteins that are often involved in promoting cell growth, hematopoiesis, and immune responses. There are blurred divisions between cytokines and hormones. For example, a growth hormone is often also considered a cytokine.
Cytokine receptors typically exist as one unit or as monomers. Cytokine binding to the receptor causes a conformational change that enables formation of homodimers (if the receptors are identical). If two or more different types of receptors dimerize this is referred to a receptor heterodimerization.
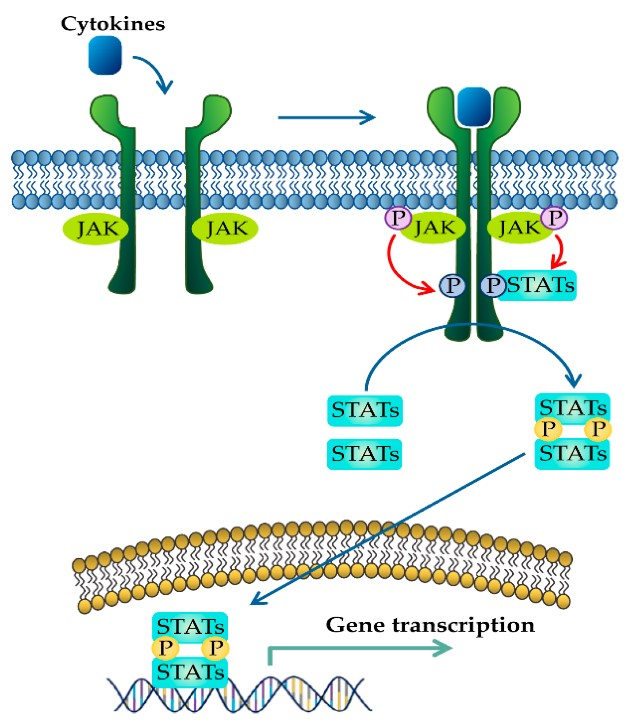
The mechanism for activation of cytokine receptors is similar to RTKs, in the manner that ligand binding results in a conformational change, bringing the monomers in close vicinity to each other. The intracellular tyrosine kinases that are associated with each receptor will be positioned in an orientation that enables them to phosphorylate the opposing kinase.
For example, cytokine receptor dimerization positions JAKs, on the receptor and they carry out trans-autophosphorylation, where JAKs phosphorylate a tyrosine on the opposite receptor. This phosphorylation activates JAK which proceeds to phosphorylate multiple sites on the C-terminal tail of each receptor.
Following phosphorylation of the cytokine receptor C-terminal tails, the phosphotyrosine serves as a binding site for different proteins including the signal transducer and activator of transcription (STAT) proteins. STAT proteins are transcription factors that contain several domains including SH2 domains. The SH2 domains serve two functions. Initially, these domains recognize and bind to the phosphotyrosine sites on the cytoplasmic C-terminal tails of the cytokine receptor. Following docking of the STAT proteins on the receptor, JAK will subsequently phosphorylate the C-terminal tail of STAT proteins (Figure 2.10). These phosphorylation events result in a conformational change, where the STAT proteins will separate from the cytokine receptor. As a result, the phoshphotyrosine of one STAT monomer will bind to the SH2 domain of another STAT protein, and vice versa, creating a homodimer. The STAT homodimer enters the nucleus and binds to regulatory DNA sequences enabling for transcription of targeted genes (Erdogan et al., 2022).
SH2 domains bind phosphotyrosine containing motifs. Specific mutations in the SH2 domain can lead to distortion of the secondary structure of the protein. Some of these distortions can negatively affect the protein structure which leads to loss-of-function. For example, a mutation K591E in the STAT3 SH2 domain results in reduced STAT3 activity since the positively charged lysine chain is replaced by a negatively charged glutamic acid (de Araujo, Orlova, et al., 2019). Since this residue side chain directly coordinates the negatively charged phospho-tyrosine, it leads to electrostatic repulsion and reduced-activity. In a similar manner, a STAT5 N642H mutation results in gain-of-function activity of the protein and this mutant is found in multiple blood cancers (de Araujo, Erdogan, et al., 2019; de Araujo et al., 2020). This mutation directly increases the affinity of the SH2 domain for phosphotyrosine via coordinating the phosphate group, which ultimately leads to tighter interactions with the cytokine receptor as well as increased lifetime of the STAT dimer and prolonging transcriptional activity.
a. STAT proteins diffuse through the nuclear pore complex
b. STAT proteins require the aid of importins
c. STATs are unable to enter the nucleus
The nucleus possesses nuclear pore complexes, which are intercalated within the nuclear membrane enabling for diffusion of small molecules (less than 60 kDa). STATs require the aid of nuclear transport receptors – importins to mediate entry in the nucleus (Figure 2.11) (Reich, 2013).
In contrast to the RTKs, Ser/Thr kinase receptors (sometimes referred to as RSTK) are members of the enzyme-coupled receptor class that expectantly phosphorylate Ser and Thr residues on target proteins.
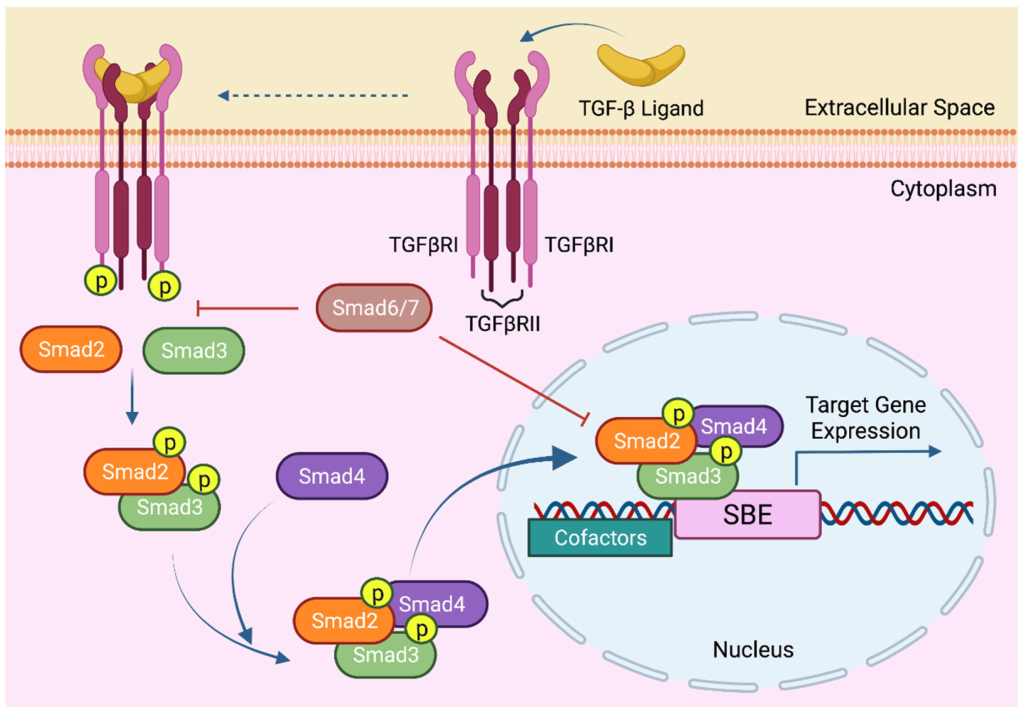
The prototypical example of RSTKs is the transforming growth factor β (TGFβ) receptors which can exist in both a Type I and Type II form. Both types have single membrane spanning domains as well as intracellular and extracellular domains. Ligands bind to the extracellular domains of Type II homodimer receptors followed by heterodimerization with Type I receptors to form a heterotetrameric complex. The intracellular ser/thr kinase domain carries out transphosphorylation. Additional modifications or variations in the C-terminal portion enable for additional layers of regulation.
While both receptors possess Ser/Thr kinase domains, Type I possess an additional ‘GS domain’ (Gomperts et al., 2009).This domain is a Gly-Ser rich domain which can also be phosphorylated. Type II receptors possess a Cys rich extracellular domain.
TGFβRII exists as a homodimer and the ligand (TGFβ) binds to the receptor (TGFβRII) resulting in a conformational change. TGFβRII binds to TGFβRI which enables serine phosphorylation. Specifically, TGFβII phosphorylates Ser rich region of the GS domain receptor of TGFβI (Tzavlaki & Moustakas, 2020). A chaperone protein (FK506 binding protein of 12 kilodalton – FKBP12) is also associated with TGFβRI. Ser phosphorylation-driven conformational changes and concurrent release of FKBP12 lead to activation of TGFβRI. At this stage, an active tetrameric receptor complex is assembled composed of two type II TGFβ receptors and two type II TGFβ receptors (Figure 2.12). Ligand binding by molecules such as TGF-β, activin, nodal, can lead to TGFβ association with Smad2 (Suppressor of Mothers against Decapentaplegic – Smad) and Smad3, while ligands such as bone morphogenetic protein (BMP) can recruit Smad1, 5, 8, or 9. In either case, Smad binding is also mediated by a membrane associated protein called SARA and the Smad protein will be phosphorylated by TGF-β. Phosphorylation of Smads results in dissociation of the phospho-Smad and SARA. The phospho-Smad forms a complex with another protein – Smad4 (Figure 2.12). Together, the phosphorylated Smad-Smad4 complex enters the nucleus to regulate gene expression.
Smads refer to a group of proteins which are transcription factors involved in regulating cell growth. Smads serve as a link between the extracellular signal (TGFβ) and a response in the nucleus. There are three members of the Smad family involved in the TGFβ pathway:
i. R-Smads (includes Smad1, 2 ,3, 5, 8/9)
ii. Co-Smads (common-mediator Smads and includes Smad4)
iii. I-Smads (inhibitory Smads which suppress the action of R-Smads in the nucleus and includes Smad6, 7)
a. In the presence of the ligand TGFβ, Smad3 is transported to the nucleus.
b. In the presence of the ligand TGFβ, Smad3 remains in the cytoplasm.
The N-terminal portion of R-Smad contains domains including a “Mad-homology 1” (MH1) domain. Part of the MH1 domain includes a NLS (Hill, 2009; Xiao et al., 2000). The nuclear-localization signal (NLS) is exposed upon phosphorylation. Proteins targeted to the nucleus possess a NLS at the N-terminus, and a mutant version of Smad3 that possesses a nonfunctional NLS will remain in the cytoplasm.
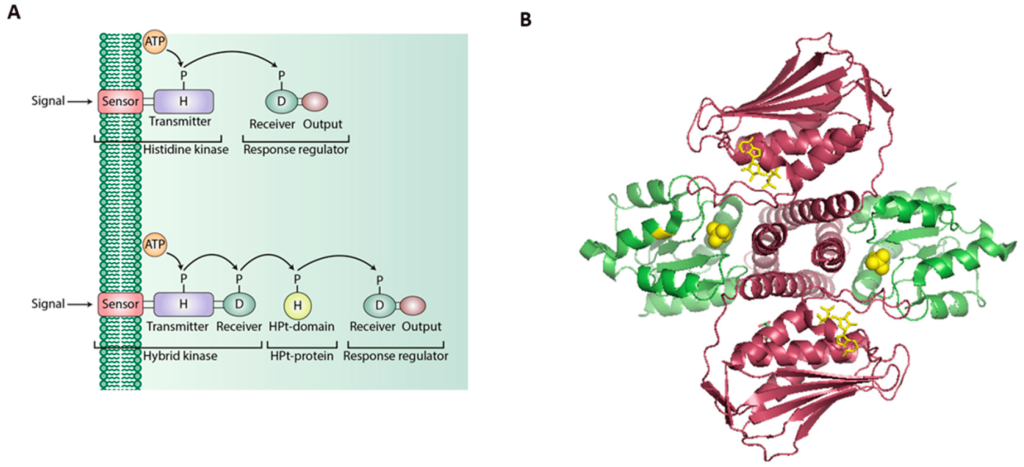
Histidine kinases transmembrane proteins dimerize upon activation. The extracellular portion contains ligand binding domains while the cytoplasmic region contains a DHp domain (Dimerization and histidine-phosphotransfer domain) along with the catalytic domain (Bhate et al., 2015; Buschiazzo & Trajtenberg, 2019). The DHp domains harbour an H box region containing the conserved His which is phosphorylated at the imidazole group (Figure 2.13). This phosphate group on His is transferred to an aspartate (Asp) on another protein, activating the protein response regulator.
Histidine kinase associated receptors are part of a ‘two-component’ cell signalling system.
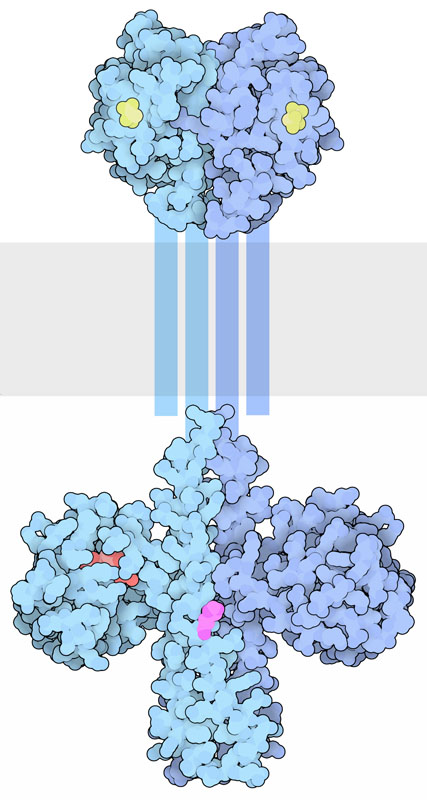
The two-component system is made up of two protein elements:
I. Sensor (containing an input domain [for ligand binding] and transmitter domain [containing the conserved His and ATP-binding/kinase domain]
II. Response regulator (containing a receiver domain with the conserved Asp and the effector or output domain)
This system is particularly important in bacteria for responding to extracellular events and mediates chemotaxis.
This system is referred to as a phosphotransfer pathway. The histidine kinase is the sensor, while the effector protein as a regulator (Figure 2.14). Upon ligand binding, the receptor (i.e. the sensor) is then activated, followed by trans-phosphorylation of a conserved His residue. The response regulator will subsequently catalyze the transfer of the phosphate from the sensor onto a specific Asp (in the receiver domain of the response regulator) (Papon & Stock, 2019). Asp phosphorylation activates the effector protein, which leads to localization to the DNA and upregulation of transcription. In more
Consistent with other receptors such as RTKs, receptor guanylyl cyclases possess both extracellular and intracellular regions flanking a transmembrane domain. The transmembrane domain consists of an alpha helical segment, while the N-terminal extracellular region contains a ligand binding domain, and the C-terminal region contains a catalytic guanylyl cyclase domain. Guanylyl cyclase enzymatic activity converts GTP into a cyclized version – guanosine 3’,5’-cyclic monophosphate (cGMP) (Figure 2.15).
a. Production of cGMP would decrease
b. Production of cGMP would increase
c. No effect on cGMP production
The enzyme phosphodiesterase catalyzes conversion of cGMP to 5’-GMP. The nucleotide cGMP is an example of a secondary messenger (Figure 2.15). The levels of cGMP decrease in the presence of phosphodiesterase.
Note, cGMP forms a complex with cGMP-dependent protein kinase G (PKG). PKG is activated and proceeds to phosphorylate target proteins potentially on Ser/Thr residues.
Similar to their counterparts (RTKs), the receptor-like protein tyrosine phosphatases are embedded within the plasma membrane with additional domains in the extracellular environment and in the cytosol. The extracellular region is variable, containing sequences similar to immunoglobulin-like motifs, carbonic anhydrase domains, or fibronectin type III-like domains (Xu & Fisher, 2012). The protein tyrosine phosphatase (PTP) domain is found in the C-terminal region and situated in the cytosol. There may be one or two copies of the PTP domain, which include a key Cys residue, essential for dephosphorylation, and the PTP domain proximal to the membrane is catalytically active, with the second domain demonstrating regulatory roles.
Broadly, receptor-like tyrosine phosphatases carry out dephosphorylation of tyrosine residues on target proteins (e.g. receptors with phosphotyrosine sites) which opposes the effects of kinases. For example, the MAPK pathway demonstrates the balancing act between kinases and phosphatases. Protein tyrosine phosphatase receptor type R (PTPRR) binds Erk (a MAPK). Erk is phosphorylated on both Tyr and Thr, resulting in activation of the kinase. PTPRR can counteract the kinase activity of Erk, by removing the phosphate groups (Xu & Fisher, 2012) which modulates Erk kinase activity.
a. Receptor-like tyrosine phosphatase activity would be inhibited.
b. Receptor-like tyrosine phosphatase activity would be activated.
Under oxidizing conditions, if the active site Cys is converted to sulphenic acid it will no longer be able to act as a strong nucleophile and remove the phosphate group (Xu & Fisher, 2012). It is predicted that the activity of Receptor-like tyrosine phosphatases would be inhibited.
Alberts, Bruce. (2002). Molecular biology of the cell. In Molecular biology of the cell (4th ed.). Garland Science. https://www.ncbi.nlm.nih.gov/books/NBK26822/
Bajusz, D., Pándy-Szekeres, G., Takács, Á., de Araujo, E. D., Erdogan, F., Neubauer, H. A., Meneksedag-Erol,raujo, E. D., & Keserű, G. M. (2023). SH2db, an information system for the SH2 domain. Nucleic Acids Research, 51(W1), W542–W552. https://doi.org/10.1093/nar/gkad420
Bhate, M. P., Molnar, K. S., Goulian, M., & DeGrado, W. F. (2015). Signal Transduction in Histidine Kinases: Insights from New Structures. Structure, 23(6), 981–994. https://doi.org/10.1016/j.str.2015.04.002
Bhagirath, A. Y., Li, Y., Patidar, R., Yerex, K., Ma, X., Kumar, A., & Duan, K. (2019). Two Component Regulatory Systems and Antibiotic Resistance in Gram-Negative Pathogens. International Journal of Molecular Sciences, 20(7). https://doi.org/10.3390/ijms20071781
Buday, L., Egan, S. E., Rodriguez Viciana, P., Cantrell, D. A., & Downward, J. (1994). A complex of Grb2 adaptor protein, Sos exchange factor, and a 36-kDa membrane-bound tyrosine phosphoprotein is implicated in ras activation in T cells. Journal of Biological Chemistry, 269(12), 9019–9023. https://doi.org/10.1016/S0021-9258(17)37070-9
Bulavin, D. V, Saito, S., Hollander, M. C., Sakaguchi, K., Anderson, C. W., Appella, E., & Fornace, A. J. (1999). Phosphorylation of human p53 by p38 kinase coordinates N‐terminal phosphorylation and apoptosis in response to UV radiation. The EMBO Journal, 18(23), 6845–6854. https://doi.org/10.1093/emboj/18.23.6845
Buschiazzo, A., & Trajtenberg, F. (2019). Two-Component Sensing and Regulation: How Do Histidine Kinases Talk with Response Regulators at the Molecular Level? Annual Review of Microbiology, 73(1), 507–528. https://doi.org/10.1146/annurev-micro-091018-054627
de Araujo, E. D., Erdogan, F., Neubauer, H. A., Meneksedag-Erol, D., Manaswiyoungkul, P., Eram, M. S., Seo, H. S., Qadree, A. K., Israelian, J., Orlova, A., Suske, T., Pham, H. T. T., Boersma, A., Tangermann, S., Kenner, L., Rülicke, T., Dong, A., Ravichandran, M., Brown, P. J., … Gunning, P. T. (2019). Structural and functional consequences of the STAT5BN642H driver mutation. Nature Communications, 10(1). https://doi.org/10.1038/s41467-019-10422-7
de Araujo, E. D., Keserű, G. M., Gunning, P. T., & Moriggl, R. (2020).Targeting STAT3 and STAT5 in Cancer. Cancers, 12(8). https://doi.org/10.3390/cancers12082002
de Araujo, E. D., Orlova, A., Neubauer, H. A., Bajusz, D., Seo, H.-S., Dhe-Paganon, S., Keserű, G. M., Moriggl, R., & Gunning, P. T. (2019). Structural Implications of STAT3 and STAT5 SH2 Domain Mutations. Cancers, 11(11). https://doi.org/10.3390/cancers11111757
Erdogan, F., Radu, T. B., Orlova, A., Qadree, A. K., de Araujo, E. D., Israelian, J., Valent, P., Mustjoki, S. M., Herling, M., Moriggl, R., & Gunning, P. T. (2022). JAK-STAT core cancer pathway: An integrative cancer interactome analysis. Journal of Cellular and Molecular Medicine, 26(7), 2049–2062. https://doi.org/10.1111/jcmm.17228
Gomperts, B. D., Kramer, I. M., & Tatham, P. E. R. (2009). Signal transduction . In Signal transduction (2nd ed.). Elsevier/Academic Press
Gungor, M. Z., Uysal, M., & Senturk, S. (2022). The Bright and the Dark Side of TGF-β Signaling in Hepatocellular Carcinoma: Mechanisms, Dysregulation, and Therapeutic Implications. Cancers, 14(4). https://doi.org/10.3390/cancers14040940
Hill, C. S. (2009). Nucleocytoplasmic shuttling of Smad proteins. Cell Research, 19(1), 36–46. https://doi.org/10.1038/cr.2008.325
Ichijo, H., Nishida, E., Irie, K., ten Dijke, P., Saitoh, M., Moriguchi, T., Takagi, M., Matsumoto, K., Miyazono, K., & Gotoh, Y. (1997). Induction of Apoptosis by ASK1, a Mammalian MAPKKK That Activates SAPK/JNK and p38 Signaling Pathways. Science, 275(5296), 90–94. https://doi.org/10.1126/science.275.5296.90
Lemmon, M. A., & Schlessinger, J. (2010). Cell Signaling by Receptor Tyrosine Kinases. Cell, 141(7), 1117–1134. https://doi.org/10.1016/j.cell.2010.06.011
Meister, M., Tomasovic, A., Banning, A., & Tikkanen, R. (2013). Mitogen-Activated Protein (MAP) Kinase Scaffolding Proteins: A Recount. International Journal of Molecular Sciences, 14(3), 4854–4884. https://doi.org/10.3390/ijms14034854
Moon, S. Y., Kim, K. D., Yoo, J., Lee, J.-H., & Hwangbo, C. (2021). Phytochemicals Targeting JAK–STAT Pathways in Inflammatory Bowel Disease: Insights from Animal Models. Molecules, 26(9). https://doi.org/10.3390/molecules26092824
Morrison, D. K. (2001). KSR: a MAPK scaffold of the Ras pathway? Journal of Cell Science, 114(9), 1609–1612. https://doi.org/10.1242/jcs.114.9.1609
Morrison, D. K. (2012). MAP kinase pathways. Cold Spring Harbor Perspectives in Biology, 4(11), a011254–a011254. https://doi.org/10.1101%2Fcshperspect.a011254.
Osaki, L. H., & Gama, P. (2013). MAPKs and Signal Transduction in the Control of Gastrointestinal Epithelial Cell Proliferation and Differentiation. International Journal of Molecular Sciences, 14(5), 10143–10161. https://doi.org/10.3390/ijms140510143
Papon, N., & Stock, A. M. (2019). Two-component systems. Current Biology, 29(15), R724–R725. https://doi.org/10.1016/j.cub.2019.06.010
Pike, L. J., Eakes, A. T., & Krebs, E. G. (1986). Characterization of affinity-purified insulin receptor/kinase. Effects of dithiothreitol on receptor/kinase function. Journal of Biological Chemistry, 261(8), 3782–3789. https://doi.org/10.1016/S0021-9258(17)35716-2
Reich, N. C. (2013). STATs get their move on. JAK-STAT, 2(4), e27080. https://doi.org/10.4161/jkst.27080
Robinson, D. R., Wu, Y.-M., & Lin, S.-F. (2000). The protein tyrosine kinase family of the human genome: Tyrosine Kinases. Oncogene, 19(49), 5548–5557. https://www.nature.com/articles/1203957
S. Corbalan-Garcia S.-S. Yang, K. R. D., & Bar-Sagi, D. (1996).Identification of the Mitogen-Activated Protein Kinase Phosphorylation Sites on Human Sosl That Regulate Interaction with Grb2. Molecular and Cellular Biology, 16(10), 5674–5682. https://doi.org/10.1128/MCB.16.10.5674
Sayem, A. S. M., Arya, A., Karimian, H., Krishnasamy, N., Ashok Hasamnis, A., & Hossain, C. F. (2018). Action of Phytochemicals on Insulin Signaling Pathways Accelerating Glucose Transporter (GLUT4) Protein Translocation. Molecules, 23(2). https://doi.org/10.3390/molecules23020258
Ségaliny, A. I., Tellez-Gabriel, M., Heymann, M.-F., & Heymann, D. (2015). Receptor tyrosine kinases: Characterisation, mechanism of action and therapeutic interests for bone cancers. Journal of Bone Oncology, 4(1), 1–12. https://doi.org/10.1016/j.jbo.2015.01.001
Tato, I., Bartrons, R., Ventura, F., & Rosa, J. L. (2011). Amino Acids Activate Mammalian Target of Rapamycin Complex 2 (mTORC2) via PI3K/Akt Signaling. The Journal of Biological Chemistry, 286(8), 6128–6142. https://doi.org/10.1074/jbc.m110.166991
Waksman, G., Shoelson, S. E., Pant, N., Cowburn, D., & Kuriyan, J. (1993). Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell, 72(5), 779–790. https://doi.org/10.1016/0092-8674(93)90405-F
Xiao, Z., Liu, X., Henis, Y. I., & Lodish, H. F. (2000). A Distinct Nuclear Localization Signal in the N Terminus of Smad 3 Determines Its Ligand-Induced Nuclear Translocation. Proceedings of the National Academy of Sciences – PNAS, 97(14), 7853–7858. https://doi.org/10.1073/pnas.97.14.7853
Xu, Y., & Fisher, G. J. (2012). Receptor type protein tyrosine phosphatases (RPTPs) – roles in signal transduction and human disease. Journal of Cell Communication and Signaling, 6(3), 125–138. https://doi.org/10.1007/s12079-012-0171-5
Yoon, M.-S. (2017). The Role of Mammalian Target of Rapamycin (mTOR) in Insulin Signaling. Nutrients, 9(11), 1176-. https://doi.org/10.3390/nu9111176
Zheng, C. F., & Guan, K. L. (1993). Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. The Journal of Biological Chemistry, 268(32), 23933–23939. https://www.jbc.org/article/S0021-9258(20)80474-8/pdf
Zhou, Y., Prakash, P., Gorfe, A. A., & Hancock, J. F. (2018). Ras and the Plasma Membrane: A Complicated Relationship. Cold Spring Harbor Perspectives in Medicine, 8(10). https://doi.org/10.1101/cshperspect.a031831
3
GPCRs are a group of receptors that play key physiological roles in biological processes, responding to cues such as aromatic compounds, neurotransmitters, hormones, and even light. These stimuli are relayed as a response to the interior of the cell. In contrast to RTKs, GPCRs activate G proteins (guanine-nucleotide binding proteins) which also forms the basis for their name. This module will focus on structural and functional aspects of GPCRs.
Upon successful completion of this chapter, you will be able to:
3.3.1 Describe the canonical structure of GPCRs.
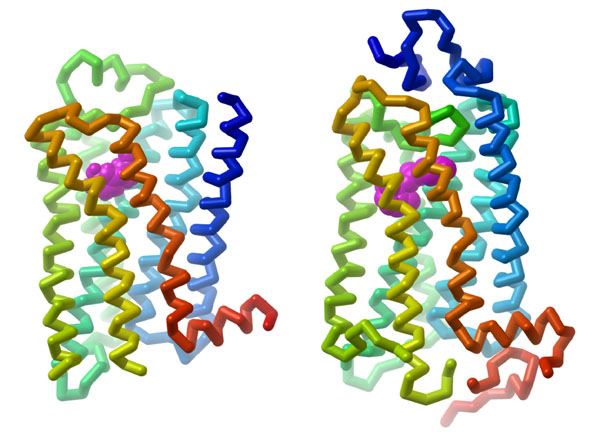
GPCRs are integral proteins (similar to enzyme coupled receptors or RTKs), that are embedded within the plasma membrane. The classical GPCR structure consists of seven transmembrane α-helices (numbered H1-H7) (Figure 3.1). The helical portions are connected by extracellular or intracellular loops. Ligands can engage with a GPCR by binding to the extracellular portion (located near at the N-terminus). The C-terminus (carboxyl portion) is situated in the cytosol and also contains an additional α-helix (H8). Importantly, GPCRs also associate with a G-protein binding domain in the cytosolic region.
a. Treatments with lipids and detergents (e.g. sodium dodecyl sulfate – SDS).
b. Incubation with low salt concentration solution (e.g. sodium chloride).
c. Increasing the pH of the buffer.
Since GPCRs contain several transmembrane helices, they require a hydrophobic environment in order to preserve their structural integrity. Due to the nature of the proteins (tightly associated with the membrane), treatment with detergents or lipid bicelles can assist with isolation of GPCRs (Weis & Kobilka, 2018). The detergents containing hydrophobic regions can interact with both the membrane and the transmembrane helices. This helps remove the GPCR from the membrane, while still maintaining the overall structure by capturing it within a lipid containing particle.
a. X-ray crystallography
b. Fluorescence microscopy
c. Differential scanning calorimetry
X-ray crystallography is a common biophysical technique to reveal protein structure information with atomic-level resolution. This technique uses X-rays that are directed in a focused beam towards a protein ‘crystal’. The X-rays will be diffracted as they pass through the crystal and produce a specific diffraction pattern (or shadow). This diffraction pattern can be mathematically deconvoluted to solve for the position of the different atoms in the crystal, which provides the three-dimensional structure of the protein.
A protein will ‘crystalize’ when water is removed from the protein sample in a very specific manner. Experimentally, this can be extremely challenging but it is a powerful technique that provides high resolution images of the atoms of the protein. Each protein will produce an unique crystal that is dependent on its three-dimensional structure.
There are a diverse number of ligands that can activate GPCRs including specific peptides, nucleotides, and photons. However, GPCRs also possess endogenous activity, albeit at a lower level compared to the presence of a ligand. Ligands which promote GPCR activity above the basal state are referred to as agonists (Weis & Kobilka, 2018). Some ligands can bind to a GPCR and block the response, which are referred to as antagonists.
There are more than 900 GPCRs identified in the human proteome and each one has a different ligand (Gaitonde & González-Maeso, 2017). Moreover, GPCRs represent targets for ~30% of all drugs that are available on the market (Hauser et al., 2017). Specific examples of endogenous GPCR ligands include epinephrine, norepinephrine, histamine, glucagon, calcitonin, oxytocin, and neurokinin.
Ligands such as peptides bind in close proximity to the amino terminus or extracellular loops. Conversely, small ligands bind within the transmembrane heptahelical region (Calebiro et al., 2021).
G proteins are guanine nucleotide-binding proteins that function as intermediary signalling proteins between GPCRs and secondary messengers. These proteins can bind both GTP and GDP which determines whether they are active (GTP-bound) or inactive (GDP-bound).
G proteins can be partitioned into two categories: i) small monomeric G proteins and ii) large heterotrimeric G proteins. Large heterotrimeric G proteins are composed of three different protein subunits: α (Gα), β (Gβ) and γ (Gγ). Both the α and γ subunits are connected to the membrane through lipid anchors (which are long hydrophobic tails that interact with the membrane). All three subunits exist in a trimeric complex including a bound GDP molecule at the Gα subunit. Upon activation of the GPCR (ligand binding), the GDP is exchanged for a GTP. This leads to dissociation of the Gα subunit from the Gβ and Gγ subunits. Both the α and βγ subunits can associate with additional proteins to facilitate signal transduction.
GPCRs can also associate with small G proteins. These proteins are more similar to the Gα subunits of heterotrimeric G proteins and also have GTP and GDP binding capabilities. The small G proteins are most commonly recognized as Ras GTPases.
a. Gα would be able to bind GMP·PNP however hydrolysis would not occur. Therefore, Gα would remain in the ‘on’ or active state.
b. Gα would be able to bind GMP·PNP and eventually convert to GMP. Therefore Gα would be converted in the ‘off’ or inactive state.
The α subunit of the heterotrimeric G protein possesses a guanine nucleotide binding site. Therefore, the α subunit can bind to GMP-PNP, especially considering the concentration is higher relative to GTP (Maegley et al., 1996) (Figure 3.2).
Conventionally, when the αβγ complex is bound to GDP, this results in the inactive state. The βγ subunits together stabilize the α subunit in the inactive state. When the αβγ complex is bound to GTP, this results in the active state, eventually leading to Gα dissociation.
a. Gα
b. Gβ
c. Gγ
The Gα subunit functions as a molecular switch possessing a GTPase domain.
a. GPCR
b. Gα
c. Gβ
d. Gγ
A GEF represents a guanine nucleotide exchange factor which enables exchanging GDP for GTP to promote GPCR activation. During the GPCR signalling pathway, a ligand will bind to the receptor on the extracellular surface. This triggers a conformational change that is propagated through the transmembrane helices and results in the intracellular side of the GPCR having GEF activity. Therefore, the activated GPCR helps to exchange GDP for GTP at the Gα subunit resulting in Gα activation.
During a period of stress, the body can react by releasing hormones such as adrenaline (or also referred to as epinephrine) which helps to surmount an appropriate response to external cues such as the fight or flight response.
The hormone signal (adrenaline) binds to the adrenergic receptor, causing a conformational change (Figure 3.3). The receptor associates and binds with the G protein located in the cytoplasmic space. The Gα subunit (shown in orange) is associated with GDP and binding to the activated GPCR triggers GEF activity and the GDP is replaced with GTP. The GTP binding leads to dissociation of the Gα and Gβγ (shown in yellow and green). The dissociated Gα (bound to GTP) interacts with an effector, such as a kinase to stimulate a downstream effect. In this pathway, the effector is the enzyme, adenylyl cyclase (AC), a transmembrane protein shown in blue.
Adenyl cyclase catalyzes the conversion of ATP to cAMP which is a powerful signalling molecule, called a secondary messenger (Figure 3.3). As the levels of the second messenger in the cytoplasm increase, the cAMP interacts with a number of proteins including protein kinase A (PKA). The nucleotide, cAMP activates PKA and PKA will subsequently phosphorylate a number of target proteins leading to activation of multiple proteins involved in glucose and lipid metabolism, thereby providing additional energy for the body.
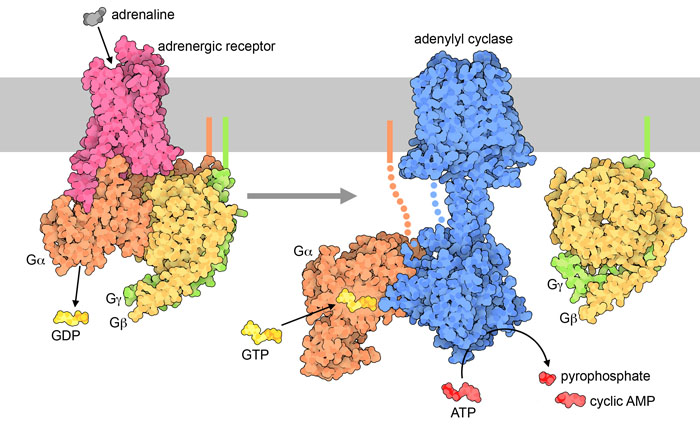
The enzyme adenylyl cyclase or adenylate cyclase is analogous to guanylyl cyclase. It catalyzes the conversion of ATP to the cyclized version (cyclic AMP – cAMP). A molecule of pyrophosphate is also released (Figure 3.3).
The extracellular signal is an example of a primary messenger, while secondary messengers are typically part of downstream signalling events. For example, the ligand that binds to a GPCR (such as epinephrine) is classified as a primary messenger. The primary messenger generally does not cross the cell membrane. Once the primary messenger binds to a target (such as a GPCR), the signal is transmitted to another molecule. In the case of epinephrine and the adrenergic receptor, cAMP is produced, and it is an example of a secondary messenger. Besides small, diffusible molecules, ions are another common example of secondary messengers. The process of converting one signal to another is referred to as signal transduction, and this facilitates responses to different stimuli. Importantly, the primary messenger can trigger the production or release of multiple secondary messenger molecules, which substantially amplifies the signal.
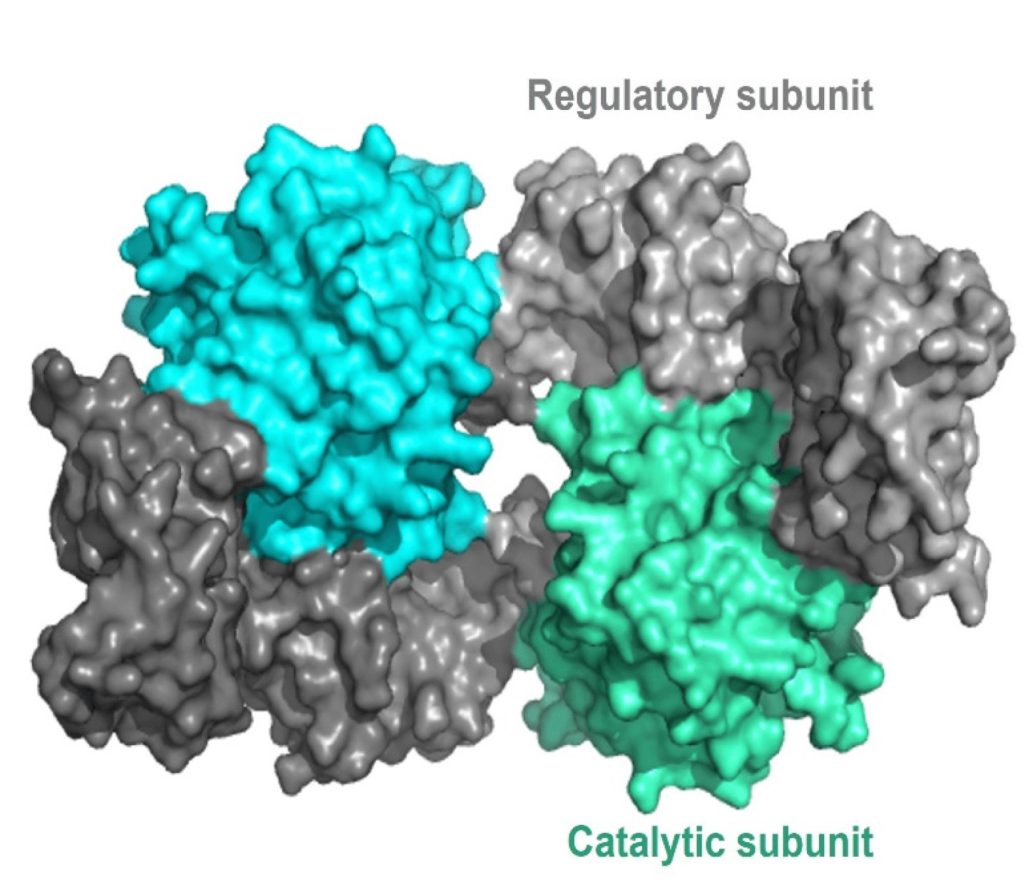
The regulatory mechanism for activation of PKA involves cAMP molecules. Structurally, PKA is a heterotetramer composed of two catalytic subunits and two regulatory subunits (Figure 3.4). The catalytic subunits harbour the active site for protein phosphorylation (and will bind ATP as well as the substrate) as well as a regulatory subunit binding domain. The regulatory subunits possess an auto-inhibitory domain (that acts as a substrate analog to bind and inhibit the kinase activity of the catalytic subunit), as well as a cAMP binding domain and a dimerization domain. Regulatory subunits exist in different isoforms, and the dimerization can be covalent via disulfide linkages or non-covalent.
In the absence (or low concentrations) of cAMP, the regulatory subunit dimer is associated with the catalytic PKA subunits, which maintains the proteins in an inactive conformation. In the presence of cAMP (or increasing concentrations), the cAMP will bind to the regulatory subunits at the cAMP binding domain, leading to allosteric modulation and dissociation of the catalytic subunits. The free PKA catalytic subunits are now capable of binding and phosphorylation downstream targets in the cell.
a. Ser
b. Thr
c. Tyr
d. a and b
PKA is a Ser/Thr kinase. However, PKA is often localized to the cell membrane through its association with a scaffold protein, A-kinase anchoring protein (AKAP) (Figure 3.5) (Welsh et al., 2023). In some cases, this introduces a layer of spatial targeting for the kinase and another mechanism for regulation.
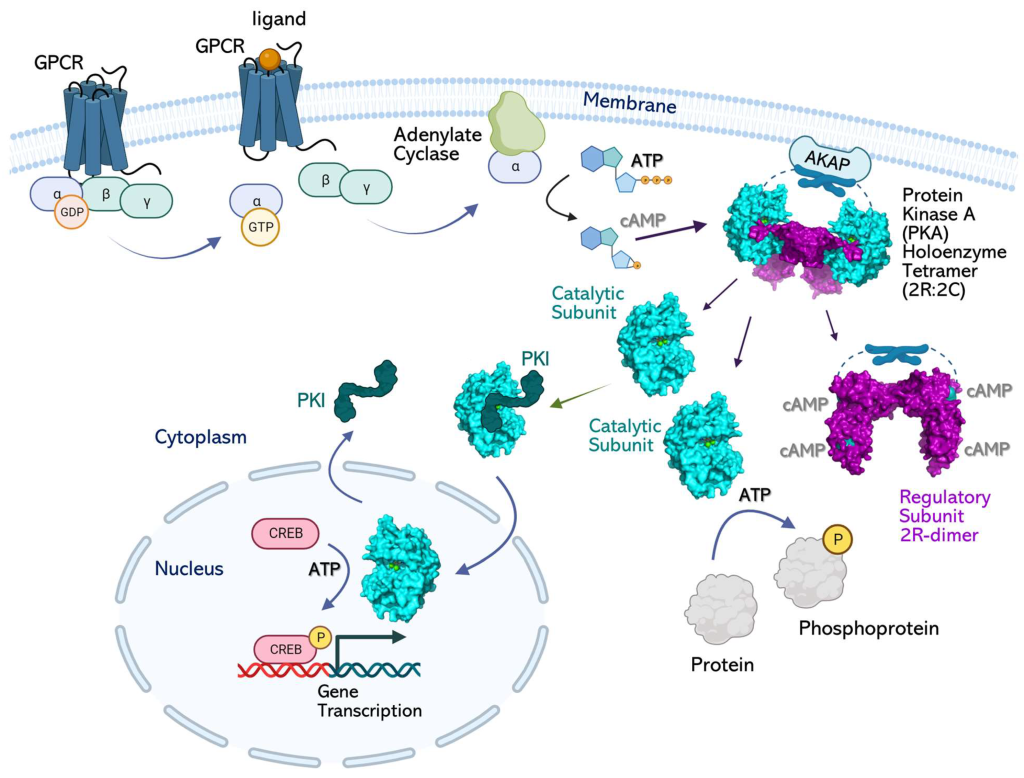
Adenyl cyclase leads to increased production of cAMP, which (as described above) activates PKA. The catalytic subunits of PKA may no longer be anchored to the membrane (if not associated with lipid anchoring proteins) and can diffuse to other regions of the cell. This includes the ability to enter the nucleus and regulate gene expression. Within the nucleus PKA phosphorylates a Ser on different transcription factors such as CRE-binding protein (CREB) (Figure 3.5). CREB can bind to another protein CBP (CREB binding protein) and the CREB/CBP complex binds to CRE (cAMP Response Element), a sequence of DNA which flanks genes under its control, including neuropeptides (e.g. somatostatin, enkephalin), tyrosine hydroxylase, and other proteins involved in circadian rhythms (Montminy et al., 1990).
This refers to proteins downstream of the pathway altered by the signal. For example, a GPCR is usually not an effector protein, but adenylyl cyclase is an effector protein. Adenylyl cyclase performs the downstream activity and carries out the “effect” of the original signal (ligand-binding event).
There are a variety of α subunits responsible for different physiological responses. For example, the αs (also referred to as Gαs) protein stimulates the enzyme adenylyl cyclase. Conversely, the inhibitor G protein α subunit – αi or Gαi protein binds and inhibits the enzyme adenylyl cyclase.
Similar to the β-adrenergic receptor (where epinephrine activates the GPCR), different ligands (such as the hormone angiotensin II) bind and activate different types of GPCRs (such as AT1 or AT2). As previously described, upon binding, activation, and GTP-exchange by the GPCR, the GTP-bound G protein α subunit dissociates from the βγ complex. The Gα subunit binds and activates the enzyme PLC or phospholipase C (the effector protein). PLC cleaves phosphatidylinositol 4,5-bisphosphate (PIP2 or PI(4,5)P2) into principal products, diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3) (Kadamur & Ross, 2013).
The fate of both of these products has substantial effects on the cell. IP3 binds to corresponding IP3-gated Ca2+ release channels on the endoplasmic reticulum (ER) membrane, leading them to open. The Ca2+ stored in the ER is released into the cytosol and can bind a number of protein targets including protein kinase C (PKC).
DAG is another product produced following PLC enzymatic action. DAG is embedded in the inner leaflet of the plasma membrane. DAG activates PKC which proceeds to phosphorylate various targets on Ser and Thr residues. Ca2+-bound PKC is capable of engaging with DAG through its DAG-binding domain (Figure 3.6).
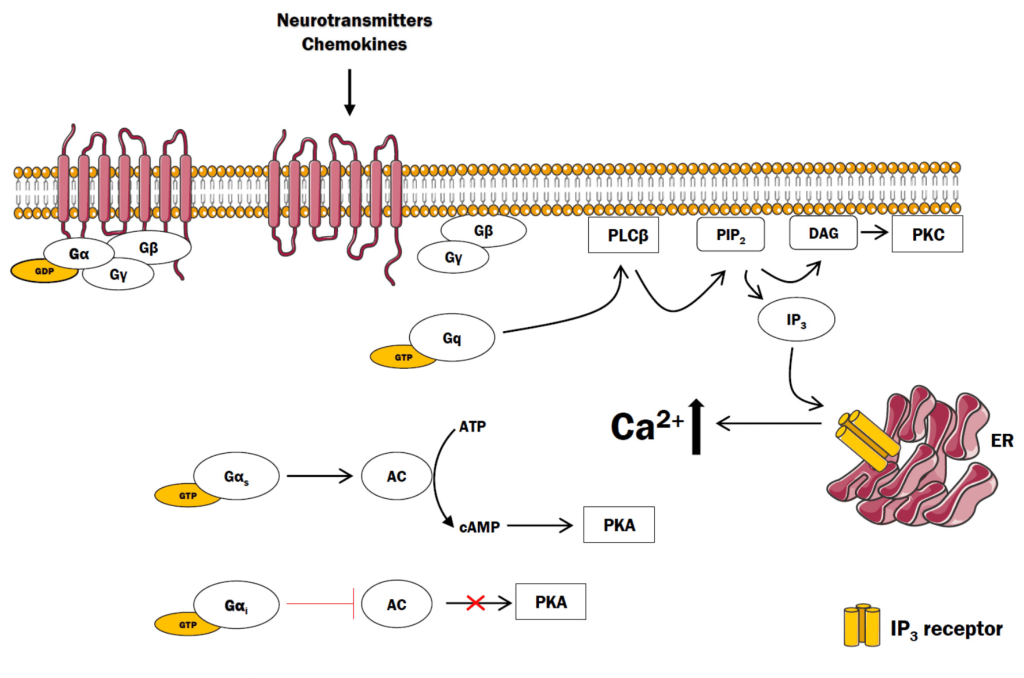
Recall, the structure of the cell membrane is a lipid bilayer is composed of an inner and outer membrane or leaflet. PIP2 is found on the inner membrane leaflet.
a. IP3
b. DAG
c. Both a and b
Both IP3 and DAG are examples of secondary messengers, where their cellular concentration increases in response to GPCR signalling pathway activation.
For simplicity, signalling pathways are often introduced as linear pathways that operate both sequentially and in isolation. However, this is rarely the case, and signalling pathways are often interconnected and lead to an intricate upregulation and downregulation of many different proteins (e.g. Figure 3.7). The activity of a single target is often governed by the consensus action of many different effector proteins. There are activators, inhibitors, chaperones, and other proteins that serve as regulators at different points along each biochemical step. This provides a high degree of regulation to precisely control the resulting physiological output from the pathway.
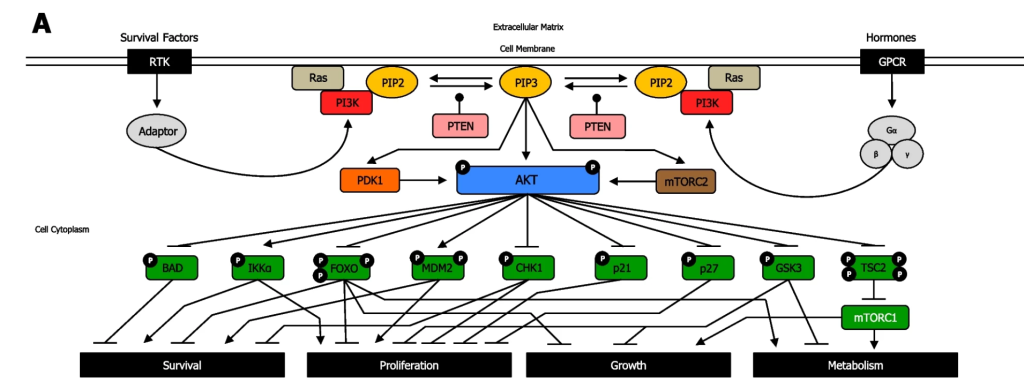
The uppercase P symbol indicates that the protein is phosphorylated.
a. Akt phosphorylates the protein BAD which results in BAD inhibition
b. Akt inhibits the BAD gene
c. Akt phosphorylates the protein BAD which results in BAD activation
d. Akt activates the BAD gene
A blunt arrowhead indicated that Akt-P inhibits the BAD protein, whereas a normal arrowhead indicates the reaction proceeds as normal. Akt phosphorylates the protein BAD which results in BAD inhibition
a. Akt Phosphorylates the protein IKKα inhibiting it
b. Akt inhibits the IKKα gene (CHUK gene)
c. Akt phosphorylates the protein IKKα activating it
d. Akt activates the IKKα gene (CHUK gene)
The arrow indicates that Akt phosphorylates the protein IKKα, thereby activating the protein.
Both RTKs and GPCRs are involved in regulating the PI3K/Akt pathway. For both receptors, the cognate ligand can bind and induce a conformational change that will impact and converge on downstream kinases – in particular the PI3K. In the case of GPCRs, it will activate PI3K, whereas in RTKs, a kinase cascade will eventually lead to activation of PI3K.
PI3K phosphorylation results in the formation of PIP3. PIP3 binds to proteins including Akt (PKB) and PDK1. The Ser/Thr kinase Akt, phosphorylates a number of downstream targets resulting in either their activation or inhibition, impacting a plethora of cellular processes. Akt phosphorylates an array of proteins such as Bcl-2-associated death promoter (BAD), forkhead box transcription factors (FOXO), Checkpoint kinase 1 (CHK1), p21, p27, Glycogen synthase kinase 3 (GSK-3) inhibiting them (Figure 3.7). Akt also phosphorylates the protein, Tuberous Sclerosis Complex 2 (TSC2), leading to inhibition, which enables mTOR complex1 to be activated. Conversely, Akt phosphorylation activates IκBα-kinase-α (IKKα) and, Mouse double minute 2 homolog (MDM2).
The phosphatase PTEN removes phosphate groups from PIP3 to form PIP2.
a. Increase
b. Decrease
c. Observe no impact on phosphorylation levels
Bisperoxovanadium, is a PTEN inhibitor (Schmid et al., 2004) and it would block the phosphatase activity of the protein. It is predicted that phosphorylation levels would increase.
Boczek, T., Mackiewicz, J., Sobolczyk, M., Wawrzyniak, J., Lisek, M., Ferenc, B., Guo, F., & Zylinska, L. (2021). The Role of G Protein-Coupled Receptors (GPCRs) and Calcium Signaling in Schizophrenia. Focus on GPCRs Activated by Neurotransmitters and Chemokines. Cells, 10(5). https://doi.org/10.3390/cells10051228
Calebiro, D., Koszegi, Z., Lanoiselée, Y., Miljus, T., & O’Brien, S. (2021). G protein-coupled receptor-G protein interactions: a single-molecule perspective. Physiological Reviews, 101(3), 857–906. https://doi.org/10.1152/physrev.00021.2020
Gaitonde, S. A., & González-Maeso, J. (2017). Contribution of heteromerization to G protein-coupled receptor function. Current Opinion in Pharmacology, 32, 23–31. https://doi.org/10.1016/j.coph.2016.10.006
Glaviano, A., Foo, A. S. C., Lam, H. Y., Yap, K. C. H., Jacot, W., Jones, R. H., Eng, H., Nair, M. G., Makvandi, P., Geoerger, B., Kulke, M. H., Baird, R. D., Prabhu, J. S., Carbone, D., Pecoraro, C., Teh, D. B. L., Sethi, G., Cavalieri, V., Lin, K. H., … Kumar, A. P. (2023). PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Molecular Cancer, 22(1), 1–138. https://doi.org/10.1186/s12943-023-01827-6
Hauser, A. S., Attwood, M. M., Rask-Andersen, M., Schiöth, H. B., & Gloriam, D. E. (2017). Trends in GPCR drug discovery: new agents, targets and indications. Nature Reviews. Drug Discovery, 16(12), 829–842. https://doi.org/10.1038/nrd.2017.178
Kadamur, G., & Ross, E. M. (2013). Mammalian phospholipase C. Annual Review of Physiology, 75(1), 127–154. https://doi.org/10.1146/annurev-physiol-030212-183750
Maegley, K. A., Admiraal, S. J., & Herschlag, D. (1996). Ras-Catalyzed Hydrolysis of GTP: A New Perspective from Model Studies. Proceedings of the National Academy of Sciences – PNAS, 93(16), 8160–8166. https://doi.org/10.1073/pnas.93.16.8160
Montminy, M. R., Gonzalez, G. A., & Yamamoto, K. K. (1990). Regulation of camp-inducible genes by creb. Trends in Neurosciences, 13(5), 184–188. https://doi.org/10.1016/0166-2236(90)90045-C
Schmid, A. C., Byrne, R. D., Vilar, R., & Woscholski, R. (2004). Bisperoxovanadium compounds are potent PTEN inhibitors. FEBS Letters, 566(1–3), 35–38. https://doi.org/10.1016/j.febslet.2004.03.102
Seok, S.-H. (2021). Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life, 11(9). https://doi.org/10.3390/life11090957
Weis, W. I., & Kobilka, B. K. (2018). The Molecular Basis of G Protein–Coupled Receptor Activation. Annual Review of Biochemistry, 87(1), 897–919. https://doi.org/10.1146/annurev-biochem-060614-033910
Welsh, C. L., Conklin, A. E., & Madan, L. K. (2023). Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A. Kinases and Phosphatases, 1(4), 265–287. https://doi.org/10.3390/kinasesphosphatases1040016
4
Nuclear receptors are transcription factors which play a role in gene expression influencing processes such as metabolism, proliferation, electrolyte balance, reproduction and inflammation (Lavery & McEwan, 2005; Mazaira GI, 2018). Due to their broad implication in a number of physiological activities, nuclear receptors are also targets for therapeutics. This unit aims to describe the mechanism of action of nuclear receptors in signalling pathways. The structural and functional aspects of nuclear receptors will be described.
Upon successful completion of this chapter, you will be able to:
a. Possess aromatic groups
b. Predominantly hydrophilic in composition
c. Predominantly hydrophobic in composition
d. Options a and b
e. Options a and c
Ligands that target nuclear receptors are distinct from ligands that target the (previously discussed) enzyme-coupled receptors or G-protein coupled receptors. This is predominantly because these ligands are relatively lipophilic and are capable of passively traversing the cell membrane and entering the cytosol to exert their effects. Ligands for enzyme-coupled receptors or GPCRs are either too large or polar and cannot cross the cell membrane.
For example, testosterone is a hormone ligand for the nuclear receptor, androgen receptor. Testosterone is comprised of a cyclopentaperhydrophenathrene ring (which is the same steroid nucleus as the common sterol, cholesterol) (Figure. 4.1). The overall structure is relatively nonpolar with the exception of two terminal oxygen atoms that cap each end of the molecule. This facilitates testosterone binding to its corresponding nuclear receptor, as the binding pocket is relatively hydrophobic with exception of the side chains of Arg752 and Thr877 making hydrogen bonding interactions with the carbonyl and hydroxyl functional groups of testosterone, respectively (de Jésus-Tran et al., 2006).
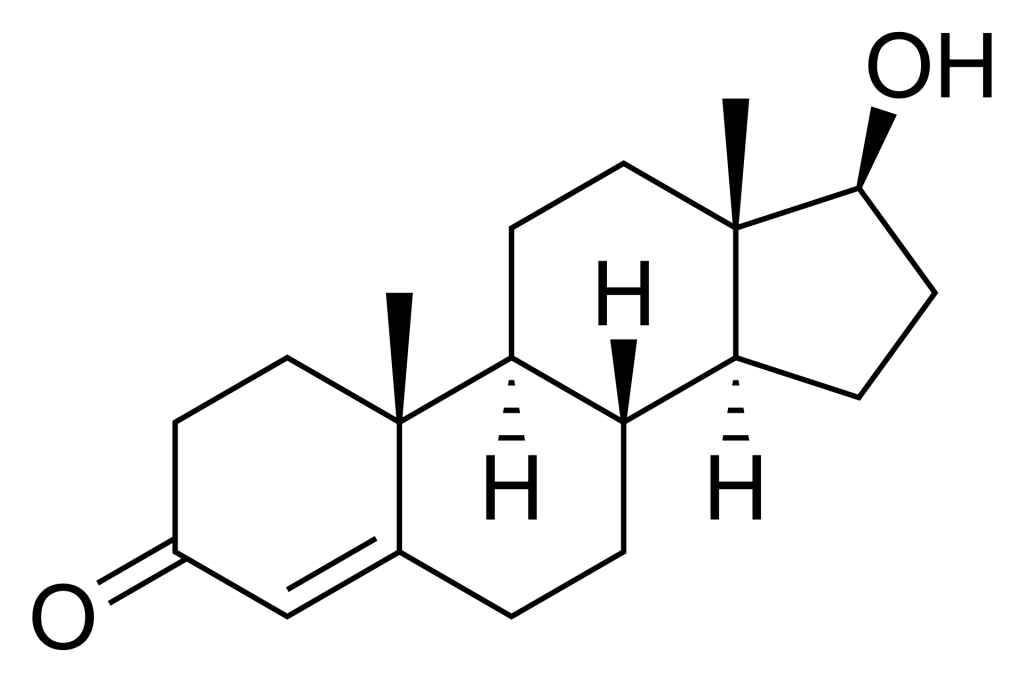
a. Nuclear receptor ligands operate in a manner similar to enzyme receptor ligands by binding in the extracellular region and exerting their effects
b. Nuclear receptor ligands cross the plasma membrane lipid bilayer, and bind receptor molecules forming a ligand-receptor complex, following entry into the nucleus
As mentioned previously, ligands that activate nuclear receptors are capable of traversing the cell membrane passively due to their lipophilic nature. Nuclear receptors are different from other receptors in that they are intracellular proteins (and not integrated into the cellular membrane). They are also known as transcription factors, because they will bind to DNA and directly facilitate transcription upon ligand binding. In fact, nuclear receptors represent the largest family of eukaryotic transcription factors (Mazaira GI, 2018).
There are 48 nuclear receptors identified in humans (Weikum et al., 2018). Over the past 50 years, these receptors have been stratified into different categories based on historical identification, sequence similarity, ligand substrate response, dimerization, and DNA-binding elements. For example, nuclear receptors were originally classified into three different types (endocrine receptors, orphan receptors, and adopted orphan receptors), and there are also classifications based on sequence similarity that have led to six sub-families (Laudet, 1997).
Conventionally, the simplest classification lists two types of nuclear receptors – called Type I and Type II receptors. Type I receptors are located in the cytoplasm, and will translocate to the nucleus following hormone binding. Androgen receptors, progesterone receptors, and glucocorticoid receptors are examples of Type I receptors. Type II receptors are found in the nucleus and are bound to DNA even in the absence of ligand. Peroxisome proliferator-activated receptors (PPAR) and retinoic acid receptor (RAR) are examples of Type II receptors.
Varying nomenclature has been employed for labelling nuclear receptors. In the most generic case, all nuclear receptors contain a region for DNA-binding and a region of ligand-binding. Based on the sequence, nuclear receptors are also labelled in regions labelled A – F (Figure 4.2). These regions contain specific domains that are important for activity. The A/B region is found at the N-terminus and is largely unstructured and contains the Activation Function-1 (AF-1). The C region contains a DNA-binding domain (DBD) which is important for binding specific sequences of DNA and initiating transcription. The D region serves as a connecting loop between the DBD and the E/F region and is sometimes called the hinge region. The E/F region is located the C-terminus and harbours the ligand-binding domain as well as the Activation Function-2 (AF-2) motif.
The DBDs enable for nuclear receptors to bind DNA at specific regions called response elements. The DBD also contains two zinc finger motifs. In addition to the different domains, nuclear receptors possess different NLSs identified in the D domain (hinge region) that facilitate cellular localization to the nucleus (Weikum et al., 2018).
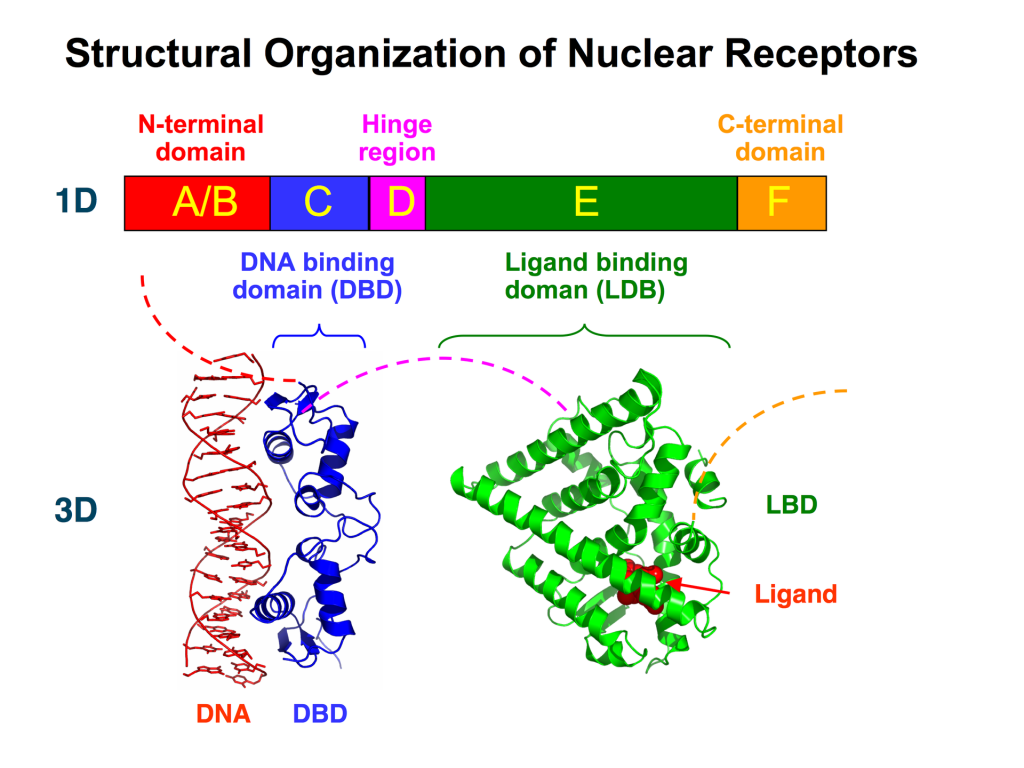
The ligands bind at the C-terminus of nuclear receptors at the E/F region or the ligand-binding domain (LBD). This triggers specific conformational changes throughout the ligand binding domain. In particular the LBD is composed of 12 α-helices along with a nonpolar pocket enabling the ligand to bind (Figure 4.2). The final helix (H12) is re-positioned to create a more condensed structure and surface pocket that enables interactions with the A/B region as well as other co-regulatory proteins. This also promotes dimerization of the nuclear receptor further reinforcing receptor activation.
For Type I receptors, ligand binding occurs (at the Ligand Binding Domain) in the cytosol following hormone diffusion across the cell membrane. The binding event triggers conformational changes that lead to dissociation of stabilizing proteins such as heat shock proteins which also enables dimerization in the cytosol. Dimerization is facilitated by both the DBD and the LBD. The AF-1 and AF-2 sites bind to other proteins that are recruited and help activate the nuclear receptor (for example SRC/p160 – p160 Steroid Receptor Coactivator family). The complex situated in the interior of the nucleus binds to hormone response elements to initiate transcription (Figure 4.3) (Feng & He, 2019).
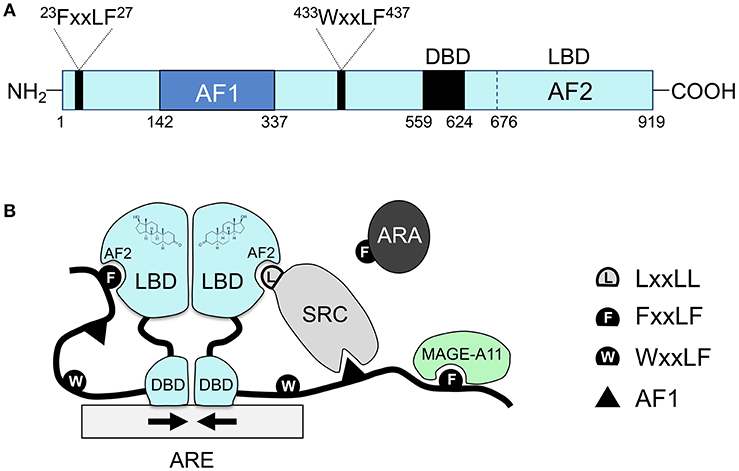
Type II receptors follow a similar pathway, except that the nuclear receptor is always bound to DNA and is usually in complex with a co-repressor protein (Sever & Glass, 2013). Binding of the ligand (e.g. hormone) liberates the release of the co-repressor, and assemblage of a co-activator and RNA polymerase which eventually leads to transcription of target proteins.
Consider a hypothetical situation, where a nuclear receptor has different Kd (dissociation constant) values for different ligands.
a. Kd = 0.5 nM
b. Kd = 5 nM
c. Kd = 50 nM
The reaction of a protein (or receptor) with a ligand is given by the equation: P + L ↔ P-L.
The association constant (Ka) can be quantitatively expressed as the concentration of the products divided by the concentration of the substrates. However, the units of this reaction are M-1 which are challenging to intuitively compare. As such, dissociations constants are commonly used to describe different reactions and provide a value that can be readily interpreted. The value of Kd represents the concentration required for half of the receptor/protein to be saturated. Therefore a lower Kd represents a more potent compound, since a lower concentration would be required engage the target. Importantly, the dissociation constant is the inverse of the association constant. Therefore, in the above example, substrate with a Kd = 0.5 nM represents the highest affinity. Importantly, different substrates/ligands can have different Kd values for different receptors. For example, estriol (an estrogen analog) has about half the Kd for estrogen receptor alpha compared to estrogen receptor beta (Dahlman-Wright et al., 2003).
a. Microtubules
b. Microfilaments
c. Intermediate filaments
Cytosolic ligand-nuclear receptor complexes are transported to the nuclear pores using microtubules as the main transport pathway (Gomperts et al., 2009).
We have described different types of nuclear receptors. In the majority of these scenarios, both the ligand and receptor have been defined (for example, the steroid cortisol binds the glucocorticoid receptor). However, there are nuclear receptors where the ligand remains unknown. These nuclear receptors are characterized as orphan receptors yet potentially still impact transcription processes either activating or repressing it (Gomperts et al., 2009). If the ligand is discovered at a later date, these receptors are updated to “adopted orphan receptors” (Li et al., 2003). For example, the PPAR receptors (alpha, gamma, and delta) are all considered adopted orphan receptors since they were originally identified as critical proteins in cell metabolism. They have been identified to respond to a number of substrates (for example, PPARα is activated by free fatty acids), and they have important pharmacological effects in a number of diseases such as hyperlipidemia and diabetes.
Kinases have been shown to phosphorylate nuclear receptors and alter their activity. For example, PKA and MAPK can phosphorylate nuclear receptors, demonstrating the cross-talk between different biochemical pathways (Adams et al., 1997; Tzagarakis-Foster & Privalsky, 1998). While multiple different sites have been identified as phosphorylation targets across all of the domains of nuclear receptors, serine residues identified in the A/B domain are common targets.
The effect of phosphorylation may promote or repress transcription. In a situation where phosphorylation enables for coactivators to bind, transcription would be upregulated. However, transcription would be inhibited if phosphorylation prevents receptor dimerization (Dongsheng Chen Paul E. Pace & Ali, 1999). For example, phosphorylation of the adrenergic receptor by PKA promotes ligand-dependent and ligand-independent activation, whereas phosphorylation of the estrogen receptor alpha by PKA can reduce dimerization and DNA binding. However, phosphorylation of different sites on the estrogen receptor alpha by other kinases (such as MAPK) can promote activation. Therefore, the effects of phosphorylation of nuclear receptor activity are complex and challenging to predict.
The are multiple different features that separate nuclear receptors from the other types of receptors. Some of the key points are listed below:
Adams, M., Reginato, M. J., Shao, D., Lazar, M. A., & Chatterjee, V. K. (1997). Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. The Journal of Biological Chemistry, 272(8), 5128–5132. https://www.jbc.org/article/S0021-9258(19)79350-8/fulltext
Dahlman-Wright, K., Koehler, K., & Gustafsson, J.-Å. (2003). Estrogen Receptor-β Structure and Function. In H. L. Henry & A. W. Norman (Eds.), Encyclopedia of Hormones (pp. 599–608). Academic Press. https://doi.org/10.1016/B0-12-341103-3/00091-7
de Jésus-Tran, K., Côté, P.-L., Cantin, L., Blanchet, J., Labrie, F., & Breton, R. (2006). Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity. Protein Science, 15(5), 987–999. https://doi.org/10.1110/ps.051905906
Dongsheng Chen Paul E. Pace, R. C. C., & Ali, S. (1999). Phosphorylation of Human Estrogen Receptor α by Protein Kinase A Regulates Dimerization. Molecular and Cellular Biology, 19(2), 1002–1015. https://doi.org/10.1128/MCB.19.2.1002
Feng, Q., & He, B. (2019). Androgen Receptor Signaling in the Development of Castration-Resistant Prostate Cancer. Frontiers in Oncology, 9. https://doi.org/10.3389/fonc.2019.00858
Gomperts, B. D., Kramer, I. M., & Tatham, P. E. R. (2009). Signal transduction . In Signal transduction (2nd ed.). Elsevier/Academic Press.
Laudet, V. (1997). Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. Journal of Molecular Endocrinology, 19(3), 207–226. https://doi.org/10.1677/jme.0.0190207
Lavery, D. N., & McEwan, I. J. (2005). Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. In Biochemical journal (Vol. 391, Issue Pt 3). Portland Press Ltd. https://doi.org/10.1042/bj20050872
Li, Y., Lambert, M. H., & Xu, H. E. (2003). Activation of Nuclear Receptors: A Perspective from Structural Genomics. Structure, 11(7), 741–746. https://doi.org/10.1016/S0969-2126(03)00133-3
Mazaira GI, Zgajnar NR, Lotufo CM, Daneri-Becerra C, Sivils JC, Soto OB, Cox MB, G. M. (2018). The Nuclear Receptor Field: A Historical Overview and Future Challenges. Nuclear Receptor Research. https://doi.org/10.11131/2018/101320
Sever, R., & Glass, C. K. (2013). Signaling by nuclear receptors. Cold Spring Harbor Perspectives in Biology, 5(3), a016709–a016709. https://doi.org/10.1101%2Fcshperspect.a016709
Tzagarakis-Foster, C., & Privalsky, M. L. (1998). Phosphorylation of Thyroid Hormone Receptors by Protein Kinase A Regulates DNA Recognition by Specific Inhibition of Receptor Monomer Binding. The Journal of Biological Chemistry, 273(18), 10926–10932. https://doi.org/10.1074/jbc.273.18.10926
Tzavlaki, K., & Moustakas, A. (2020). TGF-β signaling. Biomolecules (Basel, Switzerland), 10(3), 487-. https://doi.org/10.3390/biom10030487
Weikum, E. R., Liu, X., & Ortlund, E. A. (2018). The nuclear receptor superfamily: A structural perspective. In Protein Science (Vol. 27, Issue 11, pp. 1876–1892). https://doi.org/10.1002/pro.3496
5
Adams, M., Reginato, M. J., Shao, D., Lazar, M. A., & Chatterjee, V. K. (1997). Transcriptional activation by peroxisome proliferator-activated receptor gamma is inhibited by phosphorylation at a consensus mitogen-activated protein kinase site. The Journal of Biological Chemistry, 272(8), 5128–5132. https://www.jbc.org/article/S0021-9258(19)79350-8/fulltext
Alberts, Bruce. (2002). Molecular biology of the cell. In Molecular biology of the cell (4th ed.). Garland Science. https://www.ncbi.nlm.nih.gov/books/NBK26822/
Bajusz, D., Pándy-Szekeres, G., Takács, Á., de Araujo, E. D., & Keserű, G. M. (2023). SH2db, an information system for the SH2 domain. Nucleic Acids Research, 51(W1), W542–W552. https://doi.org/10.1093/nar/gkad420
Bhagirath, A. Y., Li, Y., Patidar, R., Yerex, K., Ma, X., Kumar, A., & Duan, K. (2019). Two Component Regulatory Systems and Antibiotic Resistance in Gram-Negative Pathogens. International Journal of Molecular Sciences, 20(7). https://doi.org/10.3390/ijms20071781
Bhate, M. P., Molnar, K. S., Goulian, M., & DeGrado, W. F. (2015). Signal Transduction in Histidine Kinases: Insights from New Structures. Structure, 23(6), 981–994. https://doi.org/10.1016/j.str.2015.04.002
Boczek, T., Mackiewicz, J., Sobolczyk, M., Wawrzyniak, J., Lisek, M., Ferenc, B., Guo, F., & Zylinska, L. (2021). The Role of G Protein-Coupled Receptors (GPCRs) and Calcium Signaling in Schizophrenia. Focus on GPCRs Activated by Neurotransmitters and Chemokines. Cells, 10(5). https://doi.org/10.3390/cells10051228
Buday, L., Egan, S. E., Rodriguez Viciana, P., Cantrell, D. A., & Downward, J. (1994). A complex of Grb2 adaptor protein, Sos exchange factor, and a 36-kDa membrane-bound tyrosine phosphoprotein is implicated in ras activation in T cells. Journal of Biological Chemistry, 269(12), 9019–9023. https://doi.org/10.1016/S0021-9258(17)37070-9
Bulavin, D. V, Saito, S., Hollander, M. C., Sakaguchi, K., Anderson, C. W., Appella, E., & Fornace, A. J. (1999). Phosphorylation of human p53 by p38 kinase coordinates N‐terminal phosphorylation and apoptosis in response to UV radiation. The EMBO Journal, 18(23), 6845–6854. https://doi.org/10.1093/emboj/18.23.6845
Buschiazzo, A., & Trajtenberg, F. (2019). Two-Component Sensing and Regulation: How Do Histidine Kinases Talk with Response Regulators at the Molecular Level? Annual Review of Microbiology, 73(1), 507–528. https://doi.org/10.1146/annurev-micro-091018-054627
Calebiro, D., Koszegi, Z., Lanoiselée, Y., Miljus, T., & O’Brien, S. (2021). G protein-coupled receptor-G protein interactions: a single-molecule perspective. Physiological Reviews, 101(3), 857–906. https://doi.org/10.1152/physrev.00021.2020
Dahlman-Wright, K., Koehler, K., & Gustafsson, J.-Å. (2003). Estrogen Receptor-β Structure and Function. In H. L. Henry & A. W. Norman (Eds.), Encyclopedia of Hormones (pp. 599–608). Academic Press. https://doi.org/10.1016/B0-12-341103-3/00091-7
de Araujo, E. D., Erdogan, F., Neubauer, H. A., Meneksedag-Erol, D., Manaswiyoungkul, P., Eram, M. S., Seo, H. S., Qadree, A. K., Israelian, J., Orlova, A., Suske, T., Pham, H. T. T., Boersma, A., Tangermann, S., Kenner, L., Rülicke, T., Dong, A., Ravichandran, M., Brown, P. J., … Gunning, P. T. (2019). Structural and functional consequences of the STAT5BN642H driver mutation. Nature Communications, 10(1). https://doi.org/10.1038/s41467-019-10422-7
de Araujo, E. D., Keserű, G. M., Gunning, P. T., & Moriggl, R. (2020).Targeting STAT3 and STAT5 in Cancer. Cancers, 12(8). https://doi.org/10.3390/cancers12082002
de Araujo, E. D., Orlova, A., Neubauer, H. A., Bajusz, D., Seo, H.-S., Dhe-Paganon, S., Keserű, G. M., Moriggl, R., & Gunning, P. T. (2019). Structural Implications of STAT3 and STAT5 SH2 Domain Mutations. Cancers, 11(11). https://doi.org/10.3390/cancers11111757
de Jésus-Tran, K., Côté, P.-L., Cantin, L., Blanchet, J., Labrie, F., & Breton, R. (2006). Comparison of crystal structures of human androgen receptor ligand-binding domain complexed with various agonists reveals molecular determinants responsible for binding affinity. Protein Science, 15(5), 987–999. https://doi.org/10.1110/ps.051905906
Dongsheng, C., Pace, P.E., R. C. C., & Ali, S. (1999). Phosphorylation of Human Estrogen Receptor α by Protein Kinase A Regulates Dimerization. Molecular and Cellular Biology, 19(2), 1002–1015. https://doi.org/10.1128/MCB.19.2.1002
Erdogan, F., Radu, T. B., Orlova, A., Qadree, A. K., de Araujo, E. D., Israelian, J., Valent, P., Mustjoki, S. M., Herling, M., Moriggl, R., & Gunning, P. T. (2022). JAK-STAT core cancer pathway: An integrative cancer interactome analysis. Journal of Cellular and Molecular Medicine, 26(7), 2049–2062. https://doi.org/10.1111/jcmm.17228
Feng, Q., & He, B. (2019). Androgen Receptor Signaling in the Development of Castration-Resistant Prostate Cancer. Frontiers in Oncology, 9. https://doi.org/10.3389/fonc.2019.00858
Gaitonde, S. A., & González-Maeso, J. (2017). Contribution of heteromerization to G protein-coupled receptor function. Current Opinion in Pharmacology, 32, 23–31. https://doi.org/10.1016/j.coph.2016.10.006
Glaviano, A., Foo, A. S. C., Lam, H. Y., Yap, K. C. H., Jacot, W., Jones, R. H., Eng, H., Nair, M. G., Makvandi, P., Geoerger, B., Kulke, M. H., Baird, R. D., Prabhu, J. S., Carbone, D., Pecoraro, C., Teh, D. B. L., Sethi, G., Cavalieri, V., Lin, K. H., … Kumar, A. P. (2023). PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Molecular Cancer, 22(1), 1–138. https://doi.org/10.1186/s12943-023-01827-6
Gomperts, B. D., Kramer, I. M., & Tatham, P. E. R. (2009). Signal transduction . In Signal transduction (2nd ed.). Elsevier/Academic Press.
Gungor, M. Z., Uysal, M., & Senturk, S. (2022). The Bright and the Dark Side of TGF-beta; Signaling in Hepatocellular Carcinoma: Mechanisms, Dysregulation, and Therapeutic Implications. Cancers, 14(4). https://doi.org/10.3390/cancers14040940
Harwood, K. H., McQuade, R. M., Jarnicki, A., & Schneider-Futschik, E. K. (2021). Anti-Inflammatory Influences of Cystic Fibrosis Transmembrane Conductance Regulator Drugs on Lung Inflammation in Cystic Fibrosis. International Journal of Molecular Sciences, 22(14). https://doi.org/10.3390/ijms22147606
Hill, C. S. (2009). Nucleocytoplasmic shuttling of Smad proteins. Cell Research, 19(1), 36–46. https://doi.org/10.1038/cr.2008.325
Ichijo, H., Nishida, E., Irie, K., ten Dijke, P., Saitoh, M., Moriguchi, T., Takagi, M., Matsumoto, K., Miyazono, K., & Gotoh, Y. (1997). Induction of Apoptosis by ASK1, a Mammalian MAPKKK That Activates SAPK/JNK and p38 Signaling Pathways. Science, 275(5296), 90–94. https://doi.org/10.1126/science.275.5296.90
Kadamur, G., & Ross, E. M. (2013). Mammalian phospholipase C. Annual Review of Physiology, 75(1), 127–154. https://doi.org/10.1146/annurev-physiol-030212-183750
Kemp, B. E., Graves, D. J., Benjamini, E., & Krebs, E. G. (1977). Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. The Journal of Biological Chemistry, 252(14), 4888–4894. https://www.jbc.org/article/S0021-9258(17)40137-2/pdf
Laudet, V. (1997). Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor. Journal of Molecular Endocrinology, 19(3), 207–226. https://doi.org/10.1677/jme.0.0190207
Lavery, D. N., & McEwan, I. J. (2005). Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. In Biochemical journal (Vol. 391, Issue Pt 3). Portland Press Ltd. https://doi.org/10.1042/bj20050872
Lemmon, M. A., & Schlessinger, J. (2010). Cell Signaling by Receptor Tyrosine Kinases. Cell, 141(7), 1117–1134. https://doi.org/10.1016/j.cell.2010.06.011
Li, Y., Lambert, M. H., & Xu, H. E. (2003). Activation of Nuclear Receptors: A Perspective from Structural Genomics. Structure, 11(7), 741–746. https://doi.org/10.1016/S0969-2126(03)00133-3
Liu, Y., & Chance, M. R. (2014). Integrating phosphoproteomics in systems biology. Computational and Structural Biotechnology Journal, 10(17), 90–97. https://doi.org/10.1016%2Fj.csbj.2014.07.003
Maegley, K. A., Admiraal, S. J., & Herschlag, D. (1996). Ras-Catalyzed Hydrolysis of GTP: A New Perspective from Model Studies. Proceedings of the National Academy of Sciences – PNAS, 93(16), 8160–8166. https://doi.org/10.1073/pnas.93.16.8160
Manning, G., Whyte, D. B., Martinez, R., Hunter, T., & Sudarsanam, S. (2002). The Protein Kinase Complement of the Human Genome. Science, 298(5600), 1912–1934. https://doi.org/10.1126/science.1075762
Mazaira GI, Zgajnar NR, Lotufo CM, Daneri-Becerra C, Sivils JC, Soto OB, Cox MB, G. M. (2018). The Nuclear Receptor Field: A Historical Overview and Future Challenges. Nuclear Receptor Research. https://doi.org/10.11131/2018/101320
Meister, M., Tomasovic, A., Banning, A., & Tikkanen, R. (2013). Mitogen-Activated Protein (MAP) Kinase Scaffolding Proteins: A Recount. International Journal of Molecular Sciences, 14(3), 4854–4884. https://doi.org/10.3390/ijms14034854
Montminy, M. R., Gonzalez, G. A., & Yamamoto, K. K. (1990). Regulation of camp-inducible genes by creb. Trends in Neurosciences, 13(5), 184–188. https://doi.org/10.1016/0166-2236(90)90045-C
Moon, S. Y., Kim, K. D., Yoo, J., Lee, J.-H., & Hwangbo, C. (2021). Phytochemicals Targeting JAK–STAT Pathways in Inflammatory Bowel Disease: Insights from Animal Models. Molecules, 26(9). https://doi.org/10.3390/molecules26092824
Morrison, D. K. (2001). KSR: a MAPK scaffold of the Ras pathway? Journal of Cell Science, 114(9), 1609–1612. https://doi.org/10.1242/jcs.114.9.1609
Morrison, D. K. (2012). MAP kinase pathways. Cold Spring Harbor Perspectives in Biology, 4(11), a011254–a011254. https://doi.org/10.1101%2Fcshperspect.a011254.
Osaki, L. H., & Gama, P. (2013). MAPKs and Signal Transduction in the Control of Gastrointestinal Epithelial Cell Proliferation and Differentiation. International Journal of Molecular Sciences, 14(5), 10143–10161. https://doi.org/10.3390/ijms140510143
Papon, N., & Stock, A. M. (2019). Two-component systems. Current Biology, 29(15), R724–R725. https://doi.org/10.1016/j.cub.2019.06.010
Pike, L. J., Eakes, A. T., & Krebs, E. G. (1986). Characterization of affinity-purified insulin receptor/kinase. Effects of dithiothreitol on receptor/kinase function. Journal of Biological Chemistry, 261(8), 3782–3789. https://doi.org/10.1016/S0021-9258(17)35716-2
Qu, C. K. (2000). The SHP—2 tyrosine phosphatase: Signaling mechanisms and biological functions. Cell Research, 10(4), 279–288. https://doi.org/10.1038/sj.cr.7290055
Reich, N. C. (2013). STATs get their move on. JAK-STAT, 2(4), e27080. https://doi.org/10.4161/jkst.27080
Robinson, D. R., Wu, Y.-M., & Lin, S.-F. (2000). The protein tyrosine kinase family of the human genome: Tyrosine Kinases. Oncogene, 19(49), 5548–5557. https://doi.org/10.1038/sj.onc.1203957
S. Corbalan-Garcia S.-S. Yang, K. R. D., & Bar-Sagi, D. (1996).Identification of the Mitogen-Activated Protein Kinase Phosphorylation Sites on Human Sosl That Regulate Interaction with Grb2. Molecular and Cellular Biology, 16(10), 5674–5682. https://doi.org/10.1128/MCB.16.10.5674
Sayem, A. S. M., Arya, A., Karimian, H., Krishnasamy, N., Ashok Hasamnis, A., & Hossain, C. F. (2018). Action of Phytochemicals on Insulin Signaling Pathways Accelerating Glucose Transporter (GLUT4) Protein Translocation. Molecules, 23(2). https://doi.org/10.3390/molecules23020258
Schmid, A. C., Byrne, R. D., Vilar, R., & Woscholski, R. (2004). Bisperoxovanadium compounds are potent PTEN inhibitors. FEBS Letters, 566(1–3), 35–38. https://doi.org/10.1016/j.febslet.2004.03.102
Ségaliny, A. I., Tellez-Gabriel, M., Heymann, M.-F., & Heymann, D. (2015). Receptor tyrosine kinases: Characterisation, mechanism of action and therapeutic interests for bone cancers. Journal of Bone Oncology, 4(1), 1–12. https://doi.org/10.1016/j.jbo.2015.01.001
Seok, S.-H. (2021). Structural Insights into Protein Regulation by Phosphorylation and Substrate Recognition of Protein Kinases/Phosphatases. Life, 11(9). https://doi.org/10.3390/life11090957
Sever, R., & Glass, C. K. (2013). Signaling by nuclear receptors. Cold Spring Harbor Perspectives in Biology, 5(3), a016709–a016709. https://doi.org/10.1101%2Fcshperspect.a016709
Tato, I., Bartrons, R., Ventura, F., & Rosa, J. L. (2011). Amino Acids Activate Mammalian Target of Rapamycin Complex 2 (mTORC2) via PI3K/Akt Signaling. The Journal of Biological Chemistry, 286(8), 6128–6142. https://doi.org/10.1074/jbc.m110.166991
Tzagarakis-Foster, C., & Privalsky, M. L. (1998). Phosphorylation of Thyroid Hormone Receptors by Protein Kinase A Regulates DNA Recognition by Specific Inhibition of Receptor Monomer Binding. The Journal of Biological Chemistry, 273(18), 10926–10932. https://doi.org/10.1074/jbc.273.18.10926
Tzavlaki, K., & Moustakas, A. (2020). TGF-β signaling. Biomolecules (Basel, Switzerland), 10(3), 487. https://doi.org/10.3390/biom10030487
Waksman, G., Shoelson, S. E., Pant, N., Cowburn, D., & Kuriyan, J. (1993). Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: Crystal structures of the complexed and peptide-free forms. Cell, 72(5), 779–790. https://doi.org/10.1016/0092-8674(93)90405-F
Weikum, E. R., Liu, X., & Ortlund, E. A. (2018). The nuclear receptor superfamily: A structural perspective. In Protein Science (Vol. 27, Issue 11, pp. 1876–1892). https://doi.org/10.1002/pro.3496
Weis, W. I., & Kobilka, B. K. (2018). The Molecular Basis of G Protein–Coupled Receptor Activation. Annual Review of Biochemistry, 87(1), 897–919. https://doi.org/10.1146/annurev-biochem-060614-033910
Welsh, C. L., Conklin, A. E., & Madan, L. K. (2023). Interaction Networks Explain Holoenzyme Allostery in Protein Kinase A. Kinases and Phosphatases, 1(4), 265–287. https://doi.org/10.3390/kinasesphosphatases1040016
Xiao, Z., Liu, X., Henis, Y. I., & Lodish, H. F. (2000). A Distinct Nuclear Localization Signal in the N Terminus of Smad 3 Determines Its Ligand-Induced Nuclear Translocation. Proceedings of the National Academy of Sciences – PNAS, 97(14), 7853–7858. https://doi.org/10.1073/pnas.97.14.7853
Xu, Y., & Fisher, G. J. (2012). Receptor type protein tyrosine phosphatases (RPTPs) – roles in signal transduction and human disease. Journal of Cell Communication and Signaling, 6(3), 125–138. https://doi.org/10.1007/s12079-012-0171-5
Yoon, M.-S. (2017). The Role of Mammalian Target of Rapamycin (mTOR) in Insulin Signaling. Nutrients, 9(11), 1176-. https://doi.org/10.3390/nu9111176
Zheng, C. F., & Guan, K. L. (1993). Properties of MEKs, the kinases that phosphorylate and activate the extracellular signal-regulated kinases. The Journal of Biological Chemistry, 268(32), 23933–23939. https://www.jbc.org/article/S0021-9258(20)80474-8/pdf
Zhou, Y., Prakash, P., Gorfe, A. A., & Hancock, J. F. (2018). Ras and the Plasma Membrane: A Complicated Relationship. Cold Spring Harbor Perspectives in Medicine, 8(10). https://doi.org/10.1101/cshperspect.a031831
1
Cover Image (PDB ID: 6MBW- Structure of Transcription Factor, Credit: de Araujo et al., 2019, Jmol: an open-source Java viewer for chemical structures in 3D. http://www.jmol.org/).
Fig 1.1 (Structures of the amino acids, Ser, Thr, and Tyr, Credit: NEUROtiker, Public Domain).
Fig 1.2 (Figure 1, Credit: Seok, 2021, CC BY license).
Fig 1.3 (Figure 1, Credit: Harwood et al., 2021, CC BY license).
Fig 2.1 (Structure of ATP, Credit: NEUROtiker, Public Domain).
Fig 2.2 (Molecule of the Month: Epidermal Growth Factor, Credit: David S. Goodsell and the RCSB PDB, 2010, CC BY 4.0 license).
Fig 2.3 (Molecule of the Month: Insulin Receptor, Credit: David S. Goodsell and the RCSB PDB, 2015, CC BY 4.0 license).
Fig 2.4 (Molecule of the Month: Insulin Receptor, Credit: David S. Goodsell and the RCSB PDB, 2015, CC BY 4.0 license).
Fig 2.5 (Figure 4, Credit: Sayem et al., 2018, CC BY 4.0 license).
Fig 2.6 (A White Light Switch on a White Wall, Credit: Tara Winstead, from Pexels, Public Domain license).
Fig 2.7 (Figure 1, Credit: Meister et al., 2013, CC BY 4.0 license).
Fig 2.8 (Figure 1, Credit: Osaki & Gama, 2013, CC BY 4.0 license).
Fig 2.9 (Molecule of the Month: RAF Protein Kinases, Credit: David S. Goodsell and the RCSB PDB, 2016, CC BY 4.0 license).
Fig 2.10 (Figure 1, Credit: Moon et al., 2021, CC BY 4.0 license).
Fig 2.11 (Figure 1, Credit: Erdogan et al., 2022, CC BY 4.0 license).
Fig 2.12 (Figure 1, Credit: Gungor et al., 2022, CC BY 4.0 license).
Fig 2.13 (Molecule of the Month: Two-component Systems, Credit: David S. Goodsell and the RCSB PDB, 2015, CC BY 4.0 license).
Fig 2.14 (Figure 2, Credit: Bhagirath et al., 2019, CC BY 4.0 license).
Fig 2.15 (Structure of cGMP, Credit: NEUROtiker, Public Domain).
Fig 3.1 (Molecule of the Month: Adrenergic Receptors, Credit: David S. Goodsell and the RCSB PDB, 2008, CC BY 4.0 license).
Fig 3.2 (5′-Guanylyl imidodiphosphate, Credit: Yikrazuul, Public Domain).
Fig 3.3 (Molecule of the Month: G Proteins, Credit: David S. Goodsell and the RCSB PDB, 2004, CC BY 4.0 license).
Fig 3.4 (Figure 2, Credit: Seok, 2021, CC BY license).
Fig 3.5 (Figure 1, Credit: Welsh et al., 2023, CC BY license).
Fig 3.6 (Figure 1, Credit: Boczek et al., 2021, CC BY license).
Fig 3.7 (Figure 2, Credit: Glaviano et al., 2023, CC BY license).
Fig 4.1 (Structure of testosterone, Credit: NEUROtiker, Public Domain).
Fig 4.2 (Structural Organization of Nuclear Receptors, Credit: Boghog2, Public Domain).
2
This resource aims to follow the guidelines in Appendix A: Checklist for Accessibility of the Accessibility Toolkit – 2nd Edition.
3
This project was funded by the Government of Ontario.
The views expressed in this publication are the views of the author and do not necessarily reflect those of the Government of Ontario or the Ontario Online Learning Consortium.
