Real Analysis of Data in Psychology, Neuroscience & Behaviour Copyright © 2024 by Ali Hashemi is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, except where otherwise noted.
Real Analysis of Data in Psychology, Neuroscience & Behaviour Copyright © 2024 by Ali Hashemi is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License, except where otherwise noted.
1
Welcome to RAD in PNB! This is an open electronic resource that is designed to aid you on your journey in understanding statistics for a variety of different psychological disciplines using simulated data based on real research. After all, a fundamental aspect of research in psychology is statistical literacy.
You might have noticed that undergraduate psychology programs almost always include at least one course on using statistics in psychological research and, perhaps to your dismay, your instructor may require you to use the statistical software R to do your assignments and tests. You probably would not be surprised to hear that meaningful learning of R is not easy (or at least it is not as easy as we would like) for many students. This can have direct impacts on students being unable to apply their statistical knowledge from the course to future research opportunities. As the instructors for statistics and other psychology courses, and a combined instructing experience of over 8 years now, we certainly recognize that there is a disconnect between what happens in our classes and the way students use (or DON’T use) statistics in later research opportunities.
But FEAR NOT!
That is where this RAD in PNB open-educational resource comes in!
Here, we have worked with an interdisciplinary group of graduate students from the Department of Psychology, Neuroscience & Behaviour at McMaster University to create a rich set of research scenarios, datasets, analysis plans, practice questions, and R scripts that are directly related to recent and/or ongoing research being conducted by the faculty members whose labs you are likely to join in the coming year or two! These graduate students and content creators are becoming experts in their fields in no small part from a thorough understanding of statistics as well as a deep mastery over R.
Contributing content to the chapters on psychological statistics are Sevda Montakhaby Nodeh and Carmen Tu. In these chapters you will find questions on many psychological phenomena like perception, attention, cognition, memory, development, narrative, music, and social perceptions. Contributing content to the chapters on statistics related to neuroscience are Matin Yousefbadi and Maheshwar Panday. In these chapters you will find questions focused on understanding electroencephalograms (EEGs), magnetic resonance imaging (MRI), functional MRI (fMRI), as well as key aspects of data wrangling and managing high dimensional datasets. Finally, contributing content to the chapters on behavioural and animal behavioural research and statistics are Brendan McEwan and Sina Zarini. In these chapters you will find questions on a variety of different animal species including bedbugs, flies, frogs, lizards, and fish. We honestly could not have found a more RAD team or made this open electronic resource without their dedication and fantastic contributions!
We hope you can use this OER to…
If you’ve found yourself here from outside of the Department of Psychology, Neuroscience & Behaviour at McMaster University, you can still benefit from the diversity of research questions and analysis techniques presented here. The universality of statistics makes this OER relevant for virtually any background you come here with. So take a look because, after all, a fundamental aspect of research in psychology is statistical literacy.
Before you get started, we want to share a few remarks about this OER. First, this is the first edition and therefore is not meant to exhaustively cover the research in PNB. We do, however, hope that as the years pass, more and more work is added to fully capture the recent and ongoing work going on and in the department. Second, you can get involved with this OER! As you complete studies in your undergraduate or graduate career, we invite you to submit a representative summary of your research, data, and analysis. This is already often done with open-access research publications, so it is not a big step to transform it into an educational resource. We (Ali & Matt) or someone on the team, will be more than happy to be involved in the process of incorporating your work. In this way, this OER will always be up-to-date with recent works. And thirdly, explore this OER with an open mind. You will notice significant variation between different research fields. You will find that in different fields — and even within fields — different researchers prefer different visualization techniques. We provide a sample of what can be done, and hope it sparks enough interest in you that you can find the visualization and analysis techniques that answer your questions best.
So, have fun and stay RAD!
– Ali & Matt
I
As a cognitive researcher at the Cognition and Attention Lab at McMaster University, you are at the forefront of exploring the intricacies of proactive control within attention processes. This line of research is profoundly significant, given that the human sensory system is overwhelmed with a vast array of information at any given second, exceeding what can be processed meaningfully. The essence of research in attention is to decipher the mechanisms through which sensory input is selectively navigated and managed. This is particularly crucial in understanding how individuals anticipate and adjust in preparation for upcoming tasks or stimuli, a phenomenon especially pertinent in environments brimming with potential distractions.
In everyday life, attentional conflicts are commonplace, manifesting when goal-irrelevant information competes with goal-relevant data for attentional precedence. An example of this (one that I’m sure we are all too familiar with) is the disruption from notifications on our mobile devices, which can divert us from achieving our primary goals, such as studying or driving. From a scientific standpoint, unravelling the strategies employed by the human cognitive system to optimize the selection and sustain goal-directed behaviour represents a formidable and compelling challenge.
Your ongoing research is a direct response to this challenge, delving into how proactive control influences the ability to concentrate on task-relevant information while effectively sidelining distractions. This facet of cognitive functionality is not just a theoretical construct; it’s the very foundation of daily human behaviour and interaction.
Your study methodically assesses this dynamic by engaging participants in a task where they must identify a sequence of words under varying conditions. The first word (T1) is presented in red, followed rapidly by a second word (T2) in white. The interval between the appearance of T1 and T2, known as the stimulus onset asynchrony (SOA), serves as a critical measure in your experiment. The distinctiveness of your study emerges in the way you manipulate the selective attention demands in each trial, classified into:
By introducing informative and uninformative cues, your investigation probes the role of proactive control. Informative cues give participants a preview of the upcoming trial type, allowing them to prepare mentally for the impending challenge. Conversely, uninformative cues act as control trials and offer no insight into the trial type. The hypothesis posits that such informative cues enable participants to proactively fine-tune their attentional focus in preparation for attentional conflict, potentially enhancing performance on selection trials with informative cues compared to those with uninformative cues.
For an overview of the different trial types please refer to the figure included below.
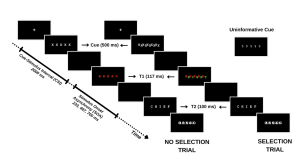
In this designed study, you are not only tackling the broader question of the role of conscious effort in attention but also contributing to a nuanced understanding of human cognitive processes, paving the way for applications that span from enhancing daily life productivity to optimizing technology interfaces for minimal cognitive disruption.
Let’s begin by running the following code in RStudio to load the required libraries. Make sure to read through the comments embedded throughout the code to understand what each line of code is doing.
# Here we create a list called "my_packages" with all of our required libraries
my_packages <- c("tidyverse", "rstatix", "readxl", "xlsx", "emmeans", "afex",
"kableExtra", "grid", "gridExtra", "superb", "ggpubr", "lsmeans")
# Checking and extracting packages that are not already installed
not_installed <- my_packages[!(my_packages %in% installed.packages()[ , "Package"])]
# Install packages that are not already installed
if(length(not_installed)) install.packages(not_installed)
# Loading the required libraries
library(tidyverse) # for data manipulation
library(rstatix) # for statistical analyses
library(readxl) # to read excel files
library(xlsx) # to create excel files
library(kableExtra) # formatting html ANOVA tables
library(superb) # production of summary stat with adjusted error bars(Cousineau, Goulet, & Harding, 2021)
library(ggpubr) # for making plots
library(grid) # for plots
library(gridExtra) # for arranging multiple ggplots for extraction
library(lsmeans) # for pairwise comparisons
Make sure to have the required dataset (“ProactiveControlCueing.xlsx“) for this exercise downloaded. Set the working directory of your current R session to the folder with the downloaded dataset. You may do this manually in R studio by clicking on the “Session” tab at the top of the screen, and then clicking on “Set Working Directory”.
If the downloaded dataset file and your R session are within the same file, you may choose the option of setting your working directory to the “source file location” (the location where your current R session is saved). If they are in different folders then click on “choose directory” option and browse for the location of the downloaded dataset.
You may also do this by running the following code:
setwd(file.choose())
Once you have set your working directory either manually or by code, in the Console below you will see the full directory of your folder as the output.
Read in the downloaded dataset as “cueingData” and complete the accompanying exercises to the best of your abilities.
# Read xlsx file
cueingData = read_excel("ProactiveControlCueing.xlsx")
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=22#h5p-1
1. Display the first few rows to understand your dataset.
head(cueingData) #Displaying the first few rows## # A tibble: 6 × 6
## ID CUE_TYPE TRIAL_TYPE SOA T1Score T2Score
## <dbl> <chr> <chr> <dbl> <dbl> <dbl>
## 1 1 INFORMATIVE NS 233 100 94.6
## 2 2 INFORMATIVE NS 233 100 97.2
## 3 3 INFORMATIVE NS 233 89.2 93.9
## 4 4 INFORMATIVE NS 233 100 91.9
## 5 5 INFORMATIVE NS 233 100 100
## 6 6 INFORMATIVE NS 233 100 97.3
2. Set up your factors and check for structure. Make sure your dependent measures are in numerical format, and that your factors and levels are set up correctly.
cueingData <- cueingData %>%
convert_as_factor(ID, CUE_TYPE, TRIAL_TYPE, SOA) #setting up factors
str(cueingData) #checking that factors and levels are set-up correctly. Checking to see that dependent measures are in numerical format.
## # A tibble: 6 × 6
## ID CUE_TYPE TRIAL_TYPE SOA T1Score T2Score
## <dbl> <chr> <chr> <dbl> <dbl> <dbl>
## 1 1 INFORMATIVE NS 233 100 94.6
## 2 2 INFORMATIVE NS 233 100 97.2
## 3 3 INFORMATIVE NS 233 89.2 93.9
## 4 4 INFORMATIVE NS 233 100 91.9
## 5 5 INFORMATIVE NS 233 100 100
## 6 6 INFORMATIVE NS 233 100 97.3
## tibble [192 × 6] (S3: tbl_df/tbl/data.frame)
## $ ID : Factor w/ 16 levels "1","2","3","4",..: 1 2 3 4 5 6 7 8 9 10 ...
## $ CUE_TYPE : Factor w/ 2 levels "INFORMATIVE",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ TRIAL_TYPE: Factor w/ 2 levels "NS","S": 1 1 1 1 1 1 1 1 1 1 ...
## $ SOA : Factor w/ 3 levels "233","467","700": 1 1 1 1 1 1 1 1 1 1 ...
## $ T1Score : num [1:192] 100 100 89.2 100 100 ...
## $ T2Score : num [1:192] 94.6 97.2 93.9 91.9 100 ...
3. Perform basic data checks for missing values and data consistency.
sum(is.na(cueingData)) # Checking for missing values in the dataset
## [1] 0
summary(cueingData) # Viewing the summary of the dataset to check for inconsistencies
## ID CUE_TYPE TRIAL_TYPE SOA T1Score
## 1 : 12 INFORMATIVE :96 NS:96 233:64 Min. : 32.43
## 2 : 12 UNINFORMATIVE:96 S :96 467:64 1st Qu.: 77.78
## 3 : 12 700:64 Median : 95.87
## 4 : 12 Mean : 86.33
## 5 : 12 3rd Qu.:100.00
## 6 : 12 Max. :100.00
## (Other):120
## T2Score
## Min. : 29.63
## 1st Qu.: 83.97
## Median : 95.76
## Mean : 87.84
## 3rd Qu.:100.00
## Max. :100.00
table(cueingData$CUE_TYPE, cueingData$TRIAL_TYPE, cueingData$SOA) #checking the number of observations per condition or combination of factors. Data is a balanced design since there is an equal number of observations per cell.## , , = 233
##
##
## NS S
## INFORMATIVE 16 16
## UNINFORMATIVE 16 16
##
## , , = 467
##
##
## NS S
## INFORMATIVE 16 16
## UNINFORMATIVE 16 16
##
## , , = 700
##
##
## NS S
## INFORMATIVE 16 16
## UNINFORMATIVE 16 16
5. The Superb library requires your dataset to be in a wide format. So convert your dataset from a long to a wide format. Save it as “cueingData.wide”.
cueingData.wide <- cueingData %>%
pivot_wider(names_from = c(TRIAL_TYPE, SOA, CUE_TYPE),
values_from = c(T1Score, T2Score) )
6. Using superbPlot() and cueingData.wide calculate the mean, and standard error of the mean (SEM) measure for T1 and T2 scores at each level of the factors. Make sure to calculate Cousineau-Morey corrected SEM values.
EXP1.T1.plot <- superbPlot(cueingData.wide,
WSFactors = c("SOA(3)", "CueType(2)", "TrialType(2)"),
variables = c("T1Score_NS_233_INFORMATIVE", "T1Score_NS_467_INFORMATIVE",
"T1Score_NS_700_INFORMATIVE", "T1Score_NS_233_UNINFORMATIVE",
"T1Score_NS_467_UNINFORMATIVE", "T1Score_NS_700_UNINFORMATIVE",
"T1Score_S_233_INFORMATIVE", "T1Score_S_467_INFORMATIVE",
"T1Score_S_700_INFORMATIVE", "T1Score_S_233_UNINFORMATIVE",
"T1Score_S_467_UNINFORMATIVE", "T1Score_S_700_UNINFORMATIVE"),
statistic = "mean",
errorbar = "SE",
adjustments = list(
purpose = "difference",
decorrelation = "CM",
popSize = 32
),
plotStyle = "line",
factorOrder = c("SOA", "CueType", "TrialType"),
lineParams = list(size=1, linetype="dashed"),
pointParams = list(size = 3))
## superb::FYI: Here is how the within-subject variables are understood:
## SOA CueType TrialType variable
## 1 1 1 T1Score_NS_233_INFORMATIVE
## 2 1 1 T1Score_NS_467_INFORMATIVE
## 3 1 1 T1Score_NS_700_INFORMATIVE
## 1 2 1 T1Score_NS_233_UNINFORMATIVE
## 2 2 1 T1Score_NS_467_UNINFORMATIVE
## 3 2 1 T1Score_NS_700_UNINFORMATIVE
## 1 1 2 T1Score_S_233_INFORMATIVE
## 2 1 2 T1Score_S_467_INFORMATIVE
## 3 1 2 T1Score_S_700_INFORMATIVE
## 1 2 2 T1Score_S_233_UNINFORMATIVE
## 2 2 2 T1Score_S_467_UNINFORMATIVE
## 3 2 2 T1Score_S_700_UNINFORMATIVE
## superb::FYI: The HyunhFeldtEpsilon measure of sphericity per group are 0.134
## superb::FYI: Some of the groups' data are not spherical. Use error bars with caution.
EXP1.T2.plot <- superbPlot(cueingData.wide,
WSFactors = c("SOA(3)", "CueType(2)", "TrialType(2)"),
variables = c("T2Score_NS_233_INFORMATIVE", "T2Score_NS_467_INFORMATIVE",
"T2Score_NS_700_INFORMATIVE", "T2Score_NS_233_UNINFORMATIVE",
"T2Score_NS_467_UNINFORMATIVE", "T2Score_NS_700_UNINFORMATIVE",
"T2Score_S_233_INFORMATIVE", "T2Score_S_467_INFORMATIVE",
"T2Score_S_700_INFORMATIVE", "T2Score_S_233_UNINFORMATIVE",
"T2Score_S_467_UNINFORMATIVE", "T2Score_S_700_UNINFORMATIVE"),
statistic = "mean",
errorbar = "SE",
adjustments = list(
purpose = "difference",
decorrelation = "CM",
popSize = 32
),
plotStyle = "line",
factorOrder = c("SOA", "CueType", "TrialType"),
lineParams = list(size=1, linetype="dashed"),
pointParams = list(size = 3)
)
## superb::FYI: Here is how the within-subject variables are understood:
## SOA CueType TrialType variable
## 1 1 1 T2Score_NS_233_INFORMATIVE
## 2 1 1 T2Score_NS_467_INFORMATIVE
## 3 1 1 T2Score_NS_700_INFORMATIVE
## 1 2 1 T2Score_NS_233_UNINFORMATIVE
## 2 2 1 T2Score_NS_467_UNINFORMATIVE
## 3 2 1 T2Score_NS_700_UNINFORMATIVE
## 1 1 2 T2Score_S_233_INFORMATIVE
## 2 1 2 T2Score_S_467_INFORMATIVE
## 3 1 2 T2Score_S_700_INFORMATIVE
## 1 2 2 T2Score_S_233_UNINFORMATIVE
## 2 2 2 T2Score_S_467_UNINFORMATIVE
## 3 2 2 T2Score_S_700_UNINFORMATIVE
## superb::FYI: The HyunhFeldtEpsilon measure of sphericity per group are 0.226
## superb::FYI: Some of the groups' data are not spherical. Use error bars with caution.
# Re-naming levels of the factors
levels(EXP1.T1.plot$data$SOA) <- c("1" = "233", "2" = "467", "3" = "700")
levels(EXP1.T2.plot$data$SOA) <- c("1" = "233", "2" = "467", "3" = "700")
levels(EXP1.T1.plot$data$TrialType) <- c("1" = "No Selection", "2" = "Selection")
levels(EXP1.T2.plot$data$TrialType) <- c("1" = "No Selection", "2" = "Selection")
levels(EXP1.T1.plot$data$CueType) <- c("1" = "Informative", "2" = "Uninformative")
levels(EXP1.T2.plot$data$CueType) <- c("1" = "Informative", "2" = "Uninformative")
7. Let’s create a beautiful printable HTML table of the summary stats for T1 and T2 Scores. This summary table can then be used in your manuscript. I suggest that you visit the following link for guides on how to create printable tables.
# Extracting summary data with CousineauMorey SEM Bars
EXP1.T1.summaryData <- data.frame(EXP1.T1.plot$data)
EXP1.T2.summaryData <- data.frame(EXP1.T2.plot$data)
# Rounding values in each column
# round(x, 1) rounds to the specified number of decimal places
EXP1.T1.summaryData$center <- round(EXP1.T1.summaryData$center,1)
EXP1.T1.summaryData$upperwidth <- round(EXP1.T1.summaryData$upperwidth,2)
EXP1.T2.summaryData$center <- round(EXP1.T2.summaryData$center,1)
EXP1.T2.summaryData$upperwidth <- round(EXP1.T2.summaryData$upperwidth,2)
# merging T1 and T2|T1 summary tables
EXP1_summarystat_results <- merge(EXP1.T1.summaryData, EXP1.T2.summaryData, by=c("TrialType","CueType","SOA"))
# Rename the column name
colnames(EXP1_summarystat_results)[colnames(EXP1_summarystat_results) == "center.x"] ="Mean"
colnames(EXP1_summarystat_results)[colnames(EXP1_summarystat_results) == "center.y"] ="Mean"
colnames(EXP1_summarystat_results)[colnames(EXP1_summarystat_results) == "upperwidth.x"] ="SEM"
colnames(EXP1_summarystat_results)[colnames(EXP1_summarystat_results) == "upperwidth.y"] ="SEM"
# deleting columns by name "lowerwidth.x" and "lowerwidth.y" in each summary table
EXP1_summarystat_results <- EXP1_summarystat_results[ , ! names(EXP1_summarystat_results) %in% c("lowerwidth.x", "lowerwidth.y")]
#removing suffixes from column names
colnames(EXP1_summarystat_results)<-gsub(".1","",colnames(EXP1_summarystat_results))
# Printable ANOVA html
EXP1_summarystat_results %>%
kbl(caption = "Summary Statistics") %>%
kable_classic(full_width = F, html_font = "Cambria", font_size = 14) %>%
add_header_above(c(" " = 3, "T1 Accuracy (%)" = 2, "T2|T1 Accuracy (%)" = 2))
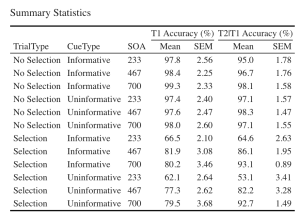
8. Use the unedited summary statistics table from Exercise 2 (EXP1.T1.summaryData and EXP1.T3.summaryData) and the ggplot() function to create separate summary line plots for T1 and T2 scores. The Line plot will visualize the relationship between SOA and the dependent measure while considering the factors of cue-type and trial-type. Your plot must have the following characteristics:
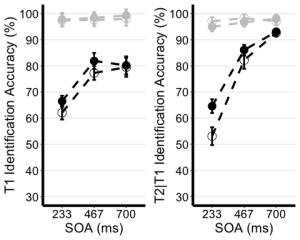
EXP1.T1.ggplotplot <- ggplot(EXP1.T1.summaryData, aes(x=SOA, y=center, color=TrialType, shape = CueType,
group=interaction(CueType, TrialType))) +
geom_point(data=filter(EXP1.T1.summaryData, CueType == "Uninformative"), shape=1, size=4.5) + # assigning shape type to level of factor
geom_point(data=filter(EXP1.T1.summaryData, CueType == "Informative"), shape=16, size=4.5) + # assigning shape type to level of factor
geom_line(linetype="dashed", linewidth=1.2) + # change line thickness and line style
scale_color_manual(values = c("gray78", "black") ) +
xlab("SOA (ms)") +
ylab("T1 Identification Accuracy (%)") +
theme_classic() + # It has no background, no bounding box.
theme(axis.line=element_line(size=1.5), # We make the axes thicker...
axis.text = element_text(size = 14, colour = "black"), # their text bigger...
axis.title = element_text(size = 16, colour = "black"), # their labels bigger...
panel.grid.major.y = element_line(), # adding horizontal grid lines
legend.position = "none") +
coord_cartesian(ylim=c(30, 100)) +
scale_y_continuous(breaks=seq(30, 100, 10)) + # Ticks from 30-100, every 10
geom_errorbar(aes(ymin=center+lowerwidth, ymax=center+upperwidth), width = 0.12, size = 1) # adding error bars from summary table
EXP1.T2.ggplotplot <- ggplot(EXP1.T2.summaryData, aes(x=SOA, y=center, color=TrialType, shape=CueType,
group=interaction(CueType, TrialType))) +
geom_point(data=filter(EXP1.T2.summaryData, CueType == "Uninformative"), shape=1, size=4.5) + # assigning shape type to level of factor
geom_point(data=filter(EXP1.T2.summaryData, CueType == "Informative"), shape=16, size=4.5) + # assigning shape type to level of factor
geom_line(linetype="dashed", linewidth=1.2) + # change line thickness and line style
scale_color_manual(values = c("gray78", "black")) +
xlab("SOA (ms)") +
ylab("T2|T1 Identification Accuracy (%)") +
theme_classic() + # It has no background, no bounding box.
theme(axis.line=element_line(size=1.5), # We make the axes thicker...
axis.text = element_text(size = 14, colour = "black"), # their text bigger...
axis.title = element_text(size = 16, colour = "black"), # their labels bigger...
panel.grid.major.y = element_line(), # adding horizontal grid lines
legend.position = "none") +
guides(fill = guide_legend(override.aes = list(shape = 16) ),
shape = guide_legend(override.aes = list(fill = "black"))) +
coord_cartesian(ylim=c(30, 100)) +
scale_y_continuous(breaks=seq(30, 100, 10)) + # Ticks from 30-100, every 10
geom_errorbar(aes(ymin=center+lowerwidth, ymax=center+upperwidth), width = 0.12, size = 1) # adding error bars from summary table
9. Use ggarrange() to display your plots together.
ggarrange(EXP1.T1.ggplotplot, EXP1.T2.ggplotplot,
nrow = 1, ncol = 2, common.legend = F,
widths = 8, heights = 5)
10. Use the data in long format (“cueingData”) and the anova_test() function, and compute a two-way ANOVA for each dependent variable, but on selection trials only. Set up cue-type and SOA as within-participants factors. (Hint: use the filter() function)
T1_2anova <- anova_test(
data = filter(cueingData, TRIAL_TYPE == "S"), dv = T1Score, wid = ID,
within = c(CUE_TYPE, SOA), detailed = TRUE, effect.size = "pes")
T2_2anova <- anova_test(
data = filter(cueingData, TRIAL_TYPE == "S"), dv = T2Score, wid = ID,
within = c(CUE_TYPE, SOA), detailed = TRUE, effect.size = "pes")
get_anova_table(T1_2anova)
## ANOVA Table (type III tests)
##
## Effect DFn DFd SSn SSd F p p<.05 pes
## 1 (Intercept) 1.00 15.00 534043.211 30705.523 260.886 6.80e-11 * 0.946
## 2 CUE_TYPE 1.00 15.00 253.665 506.454 7.513 1.50e-02 * 0.334
## 3 SOA 1.29 19.35 5091.853 2660.275 28.710 1.16e-05 * 0.657
## 4 CUE_TYPE:SOA 2.00 30.00 77.140 1061.051 1.091 3.49e-01 0.068
get_anova_table(T2_2anova)
## ANOVA Table (type III tests)
##
## Effect DFn DFd SSn SSd F p p<.05 pes
## 1 (Intercept) 1 15 593913.106 11288.706 789.169 2.19e-14 * 0.981
## 2 CUE_TYPE 1 15 661.761 426.947 23.250 2.24e-04 * 0.608
## 3 SOA 2 30 20001.563 3852.629 77.875 1.33e-12 * 0.838
## 4 CUE_TYPE:SOA 2 30 519.154 1298.144 5.999 6.00e-03 * 0.286
Exercise 5: Post-hoc tests
11. Filter for selection trials first, then group your data by SOA and use the pairwise_t_test() function to compare informative and uninformative trials at each level of SOA store and display your computation as “T2_sel_pwc”.
T2_sel_pwc <- filter(cueingData, TRIAL_TYPE == "S") %>%
group_by(SOA) %>%
pairwise_t_test(T2Score ~ CUE_TYPE, paired = TRUE, p.adjust.method = "holm", detailed = TRUE) %>%
add_significance("p.adj")
T2_sel_pwc <- get_anova_table(T2_sel_pwc)
T2_sel_pwc
## # A tibble: 3 × 16
## SOA estimate .y. group1 group2 n1 n2 statistic p df
## <fct> <dbl> <chr> <chr> <chr> <int> <int> <dbl> <dbl> <dbl>
## 1 233 11.5 T2Score INFORMATIVE UNINFO… 16 16 4.36 5.55e-4 15
## 2 467 3.89 T2Score INFORMATIVE UNINFO… 16 16 1.97 6.8 e-2 15
## 3 700 0.356 T2Score INFORMATIVE UNINFO… 16 16 0.190 8.52e-1 15
## # ℹ 6 more variables: conf.low <dbl>, conf.high <dbl>, method <chr>,
## # alternative <chr>, p.adj <dbl>, p.adj.signif <chr>
Welcome! In this assignment, we will be entering a cognition and memory lab at McMaster University. Specifically, we will be examining data from an intriguing cognitive psychology study that explores the role of repetition in recognition memory.
Most of us are familiar with the phrase ‘practice makes perfect’. This motivational idiom aligns with intuition and is confirmed by many real-world observations. Much empirical research also supports this view–repeated opportunities to encode a stimulus improve subsequent memory retrieval and perceptual identification. These observations suggest that stimulus repetition strengthens underlying memory representations.
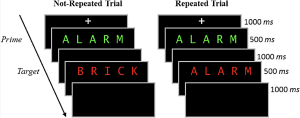
The present study focuses on a contradictory idea, that stimulus repetition can weaken memory encoding. The experiment comprised of three stages: a study phase, a distractor phase, and a surprise recognition memory test.
In the study phase participants aloud a red target word preceded by a briefly presented green prime word. On half of the trials, the prime and target were the same (repeated trials), and on the other half of the trials, the prime and target were different (not-repeated trials). In the figure below you can see an overview of the two different trial types. Following the study phase, participants engaged in a 10-minute distractor task consisting of math problems they had to solve by hand.
The final phase was a surprise recognition memory test where on each test trial they were shown a red word and asked to respond old if the word on the test was one they had previously seen at study, and new if they had never encountered the word before. Half of the trials at the test were words from the study phase and the other half were new words.
Let’s begin by running the following code to load the required libraries. Make sure to read through the comments embedded throughout the code to understand what each line of code is doing.
# Load necessary libraries
library(rstatix) #for performing basic statistical tests
library(dplyr) #for sorting data
library(readxl) #for reading excel files
library(tidyr) #for data sorting and structure
library(ggplot2) #for visualizing your data
library(plotrix) #for computing basic summary stats
Make sure to have the required dataset (“RepDecrementdataset.xlsx”) for this exercise downloaded. Set the working directory of your current R session to the folder with the downloaded dataset. You may do this manually in R studio by clicking on the “Session” tab at the top of the screen, and then clicking on “Set Working Directory”.
If the downloaded dataset file and your R session are within the same file, you may choose the option of setting your working directory to the “source file location” (the location where your current R session is saved). If they are in different folders then click on “choose directory” option and browse for the location of the downloaded dataset.
You may also do this by running the following code
setwd(file.choose())
Once you have set your working directory either manually or by code, in the Console below you will see the full directory of your folder as the output.
Read in the downloaded dataset as “MemoryData” and complete the accompanying exercises to the best of your abilities.
MemoryData <- read_excel('RepDecrementdataset.xlsx')
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=26#h5p-2
1. Display the first few rows of your dataset to familiarize yourself with its structure and contents.
head(MemoryData) #Displaying the first few rows
## # A tibble: 6 × 7
## ID Hits_NRep Hits_Rep FalseAlarms Misses_Nrep Misses_Rep CorrectRej
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 46 34 13 14 26 107
## 2 2 43 44 27 17 16 93
## 3 3 43 35 23 17 24 97
## 4 4 37 36 56 23 24 64
## 5 5 39 35 49 21 25 71
## 6 6 38 43 28 22 17 92
str(MemoryData) #Checking structure of dataset
## tibble [24 × 7] (S3: tbl_df/tbl/data.frame)
## $ ID : num [1:24] 1 2 3 4 5 6 7 8 9 10 ...
## $ Hits_NRep : num [1:24] 46 43 43 37 39 38 20 24 36 38 ...
## $ Hits_Rep : num [1:24] 34 44 35 36 35 43 11 29 43 27 ...
## $ FalseAlarms: num [1:24] 13 27 23 56 49 28 4 11 46 9 ...
## $ Misses_Nrep: num [1:24] 14 17 17 23 21 22 40 36 23 22 ...
## $ Misses_Rep : num [1:24] 26 16 24 24 25 17 49 31 17 33 ...
## $ CorrectRej : num [1:24] 107 93 97 64 71 92 116 109 74 111 ...
colnames(MemoryData)
## [1] "ID" "Hits_NRep" "Hits_Rep" "FalseAlarms" "Misses_Nrep"
## [6] "Misses_Rep" "CorrectRej"
2. Calculate Total Trials for Each Condition:
Note that if the value in “TotalNRep” and “TotalRep” is less than 60 for a participant, it indicates that certain word trials were excluded during the study phase due to issues (e.g., the participant read aloud the prime word instead of the target, leading to trial spoilage).
MemoryData <- MemoryData %>%
mutate(TotalNRep = Hits_NRep + Misses_Nrep)
MemoryData <- MemoryData %>%
mutate(TotalRep = Hits_Rep + Misses_Rep)
MemoryData <- MemoryData %>%
mutate(TotalNew = FalseAlarms + CorrectRej)
3. Transform the counts in the hits, misses, false alarms, and correct rejections columns into proportions. Do this by dividing each count by the total number of trials for the respective condition (e.g., divide hits for non-repeated trials by “TotalNRep”).
MemoryData$Hits_NRep <- (MemoryData$Hits_NRep/MemoryData$TotalNRep)
MemoryData$Misses_Nrep <- (MemoryData$Misses_Nrep/MemoryData$TotalNRep)
MemoryData$Hits_Rep <- (MemoryData$Hits_Rep/MemoryData$TotalRep)
MemoryData$Misses_Rep <- (MemoryData$Misses_Rep/MemoryData$TotalRep)
MemoryData$CorrectRej <- (MemoryData$CorrectRej/MemoryData$TotalNew)
MemoryData$FalseAlarms <- (MemoryData$FalseAlarms/MemoryData$TotalNew)
4. Once the proportions are calculated, remove the “TotalNew”, “TotalRep”, and “TotalNRep” columns from the dataset as they are no longer needed for further analysis.
MemoryData <- MemoryData[, !names(MemoryData) %in% c("TotalNew", "TotalRep","TotalNRep")]
5. Use the pivot_longer() function from the tidyr package to convert your data from wide to long format. Pivot the columns “Hits_NRep”, “Hits_Rep”, and “FalseAlarms”, setting the new column names to “Condition” and “Proportion” for the reshaped data.
long_df <- MemoryData %>%
pivot_longer(
cols = c(Hits_NRep, Hits_Rep, FalseAlarms),
names_to = "Condition",
values_to = "Proportion"
)
6. Using your long formatted dataset, group your data by ID and calculate the per-subject mean and the grand mean of the Proportions column.
data_adjusted <- long_df %>%
group_by(ID) %>%
mutate(SubjectMean = mean(Proportion, na.rm = TRUE)) %>%
ungroup() %>%
mutate(GrandMean = mean(Proportion, na.rm = TRUE)) %>%
mutate(AdjustedScore = Proportion - SubjectMean + GrandMean)
# Calculate the mean and SEM of the adjusted scores
summary_df <- data_adjusted %>%
group_by(Condition) %>%
summarize(
AdjustedMean = mean(AdjustedScore, na.rm = TRUE),
AdjustedSEM = sd(AdjustedScore, na.rm = TRUE) / sqrt(n())
)
7. Create a bar plot where the x-axis represents the Prime-target Encoding task conditions, the y-axis shows the adjusted mean proportion of old responses, and include error bars represent the adjusted SEM. Begin by setting custom colours for each condition. The colour for the bar presenting the false alarms or “New” should be “gray89”; the colour for “Hits_Nrep” or “Non-Repeated Targets” bar should be “gray39”; the colour for the “Hits_Rep” or “Repeated Targets” bar should be “darkgrey”.
# Create the bar plot with adjusted SEM error bars
ggplot(summary_df, aes(x = Condition, y = AdjustedMean, fill = Condition)) +
geom_bar(stat = "identity", position = position_dodge()) +
geom_errorbar(aes(ymin = AdjustedMean - AdjustedSEM, ymax = AdjustedMean + AdjustedSEM), width = 0.2, position = position_dodge(0.9)) +
scale_fill_manual(values = c("Hits_NRep" = "gray39", "Hits_Rep" = "darkgrey", "FalseAlarms" = "gray89"),
labels = c("Non-Repeated Targets", "Repeated Targets", "New")) +
labs(
x = "Prime Target Encoding Task",
y = "Adjusted Mean Proportion of Old Responses",
fill = "Condition"
) +
scale_y_continuous(breaks = seq(0, 1, by = 0.1), limits = c(0, 1)) +
theme_minimal(base_size = 14) +
theme(
axis.line = element_line(color = "black"),
axis.title = element_text(color = "black"),
panel.grid.major = element_line(color = "grey", size = 0.5),
panel.grid.minor = element_blank(),
legend.title = element_text(color = "black")
)
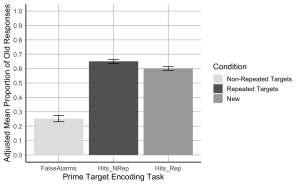
8. Using the wide formatted data file “MemoryData” conduct a two-paired sample t-test comparing the hit rate collapsed across the two repetition conditions (repeated/not-repeated) to the false alarm rate to assess participants’ ability to distinguish old from new items.
collapsed_hitdata <- MemoryData %>%
mutate(HitRate = (Hits_NRep + Hits_Rep) / 2)
# Conduct paired sample t-tests
t_test_results <- t.test(collapsed_hitdata$HitRate, collapsed_hitdata$FalseAlarms, paired = TRUE)
print(t_test_results)
##
## Paired t-test
##
## data: collapsed_hitdata$HitRate and collapsed_hitdata$FalseAlarms
## t = 11.621, df = 23, p-value = 4.179e-11
## alternative hypothesis: true mean difference is not equal to 0
## 95 percent confidence interval:
## 0.3071651 0.4401983
## sample estimates:
## mean difference
## 0.3736817
#Hit rates were higher than false alarm rates, t(23) = 11.62, p < .001.
9. Using the wide formatted data file “MemoryData” conduct a two-paired sample t-test comparing the hit rates for the not-repeated and repeated targets.
# Conduct paired sample t-tests for non-repeated vs repeated hit rates
t_test_results_hits <- t.test(collapsed_hitdata$Hits_NRep, collapsed_hitdata$Hits_Rep, paired = TRUE)
# Print the results for the hit rate comparison
print(t_test_results_hits)
##
## Paired t-test
##
## data: collapsed_hitdata$Hits_NRep and collapsed_hitdata$Hits_Rep
## t = 2.5431, df = 23, p-value = 0.01817
## alternative hypothesis: true mean difference is not equal to 0
## 95 percent confidence interval:
## 0.009071364 0.088174399
## sample estimates:
## mean difference
## 0.04862288
#Hit rates were higher for not-repeated targets than for repeated targets, t(23) = 2.54, p = .018.
You are a researcher at the Early Childhood Development Research Center. Your latest project investigates how infants respond to different combinations of face race and music emotion. In specific you are interested in whether infants associate own- and other-race faces with music of different emotional valences (happy and sad music).
Your project was completed in collaboration with your colleagues in China. While you were responsible for designing your experiment, your collaborators were responsible for recruiting participants and collecting your data.
Chinese infants (3 to 9 months old) were recruited to participate in your experiment. Each infant was randomly assigned to one of the four face-race + music conditions where they saw a series of neutral own- or other-race faces paired with happy or sad musical excerpts.
In the own-happy, infants watched six Asian face videos sequentially paired with six happy musical excerpts. In other-sad, infants watched six African face videos sequentially paired with happy musical excerpts. In general, conditions were procedurally the same, except for the face-music composition. Infant eye movements were recorded using an eye tracker.
Your goal is to determine how face race and music emotion, as well as their interaction, influence the looking behaviour of infants.
Your independent variables:
Your dependent variables:
Let’s begin by loading the required libraries and the dataset as “BabyData”. To do so download the file “infant_eye_tracking_study.csv” and run the following code. Remember to replace ‘path_to_your_downloaded_file’ with the actual path to the dataset on your system.
BabyData <- read.csv('path_to_your_downloaded_file/infant_eye_tracking_study.csv')
library(rstatix) #for performing basic statistical tests
library(dplyr) #for sorting data
library(tidyr) #for data sorting and structure
library(ggplot2) #for visualizing your data
library(readr)
library(ggpubr)
library(gridExtra)
Please complete the accompanying exercises to the best of your abilities.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=30#h5p-3
1. Display the first few rows to understand your dataset.
summary(BabyData) # Viewing the summary of the dataset to check for inconsistencies
## Age.in.Days Condition Face.Race Music.Emotion Age.Group
## 1 93 Other-Race Happy Music African happy 3
## 2 98 Other-Race Happy Music African happy 3
## 3 93 Other-Race Happy Music African happy 3
## 4 93 Other-Race Happy Music African happy 3
## 5 93 Other-Race Happy Music African happy 3
## 6 100 Other-Race Happy Music African happy 3
## Total.Looking.Time First.Face.Looking.Time Participant.ID
## 1 44.035 8.273 HJOGM7704U
## 2 18.324 6.938 JHSEG5414N
## 3 24.600 4.225 OCQFX4970K
## 4 12.919 7.537 KLDOF5559R
## 5 12.755 4.230 HHPGJ9661Y
## 6 38.777 9.351 NVCPX9518V
2. Use relocate() to re-order your columns such that your “Participant.ID” column appears as the first column in your dataset.
BabyData <- BabyData %>% relocate(Participant.ID, .before = Age.in.Days)
3. Check your data for any missing values. Remove any rows with missing or NA values from the dataset.
sum(is.na(BabyData)) # Checking for missing values in the dataset
## [1] 3
BabyData <- BabyData[!is.na(BabyData$First.Face.Looking.Time), ]
## Participant.ID Age.in.Days Condition Face.Race
## Length:193 Min. : 79.0 Length:193 Length:193
## Class :character 1st Qu.:127.0 Class :character Class :character
## Mode :character Median :185.0 Mode :character Mode :character
## Mean :189.3
## 3rd Qu.:246.0
## Max. :316.0
##
## Music.Emotion Age.Group Total.Looking.Time First.Face.Looking.Time
## Length:193 Min. :3.000 Min. : 1.654 Min. : 0.160
## Class :character 1st Qu.:3.000 1st Qu.:20.671 1st Qu.: 5.309
## Mode :character Median :6.000 Median :30.381 Median : 7.495
## Mean :6.093 Mean :29.196 Mean : 7.041
## 3rd Qu.:9.000 3rd Qu.:38.196 3rd Qu.: 9.185
## Max. :9.000 Max. :50.000 Max. :11.823
## NA's :3
4. Check your data again for any missing values and check data consistency.
sum(is.na(BabyData)) # Checking for missing values in the dataset
## [1] 0
summary(BabyData) # Viewing the summary of the dataset to check for inconsistencies
## Participant.ID Age.in.Days Condition Face.Race
## Length:193 Min. : 79.0 Length:193 Length:193
## Class :character 1st Qu.:127.0 Class :character Class :character
## Mode :character Median :185.0 Mode :character Mode :character
## Mean :189.3
## 3rd Qu.:246.0
## Max. :316.0
##
## Music.Emotion Age.Group Total.Looking.Time First.Face.Looking.Time
## Length:193 Min. :3.000 Min. : 1.654 Min. : 0.160
## Class :character 1st Qu.:3.000 1st Qu.:20.671 1st Qu.: 5.309
## Mode :character Median :6.000 Median :30.381 Median : 7.495
## Mean :6.093 Mean :29.196 Mean : 7.041
## 3rd Qu.:9.000 3rd Qu.:38.196 3rd Qu.: 9.185
## Max. :9.000 Max. :50.000 Max. :11.823
## NA's :0
5. Check for structure and ensure that your factor columns (Music.Emotion, Face.Race, and Condition) are set-up correctly.
str(BabyData)
## 'data.frame': 190 obs. of 8 variables:
## $ Participant.ID : chr "HJOGM7704U" "JHSEG5414N" "OCQFX4970K" "KLDOF5559R" ...
## $ Age.in.Days : int 93 98 93 93 93 100 93 91 98 100 ...
## $ Condition : chr "Other-Race Happy Music" "Other-Race Happy Music" "Other-Race Happy Music" "Other-Race Happy Music" ...
## $ Face.Race : chr "African" "African" "African" "African" ...
## $ Music.Emotion : chr "happy" "happy" "happy" "happy" ...
## $ Age.Group : int 3 3 3 3 3 3 3 3 3 3 ...
## $ Total.Looking.Time : num 44 18.3 24.6 12.9 12.8 ...
## $ First.Face.Looking.Time: num 8.27 6.94 4.22 7.54 4.23 ...
BabyData$Face.Race <- as.factor(BabyData$Face.Race)
BabyData$Music.Emotion <- as.factor(BabyData$Music.Emotion)
BabyData$Condition <- as.factor(BabyData$Condition)
6. Check to see if your design is balanced or unbalanced.
table(BabyData$Age.Group, BabyData$Condition) #unbalanced design
##
## Other-Race Happy Music Other-Race Sad Music Own-Race Happy Music
## 3 16 12 12
## 6 15 19 19
## 9 14 17 17
##
## Own-Race Sad Music
## 3 17
## 6 15
## 9 17
7. Conduct a multi-variable linear regression on the first face looking time as the predicted variable, with Group, face race, and their interactions as the predictors. Display the result.
lm_model1 <- lm(First.Face.Looking.Time ~ Age.Group*Face.Race, data = BabyData)
summary(lm_model1)
##
## Call:
## lm(formula = First.Face.Looking.Time ~ Age.Group * Face.Race,
## data = BabyData)
##
## Residuals:
## Min 1Q Median 3Q Max
## -7.4524 -1.4478 0.3645 2.0507 4.5670
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.50710 0.75573 7.287 8.75e-12 ***
## Age.Group 0.22815 0.11542 1.977 0.0496 *
## Face.RaceChinese -0.04233 1.05722 -0.040 0.9681
## Age.Group:Face.RaceChinese 0.05036 0.16071 0.313 0.7544
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.658 on 186 degrees of freedom
## Multiple R-squared: 0.05411, Adjusted R-squared: 0.03885
## F-statistic: 3.546 on 3 and 186 DF, p-value: 0.01564
8. Conduct a multivariable linear regression similar to the one described in the previous question. You predicted variable should be Total looking time, with Age.Group, Face.Race, Musical.Emotion, and their interactions as the predictors.
lm_model2 <- model <- lm(Total.Looking.Time ~ Age.Group * Face.Race * Music.Emotion, data = BabyData)
summary(lm_model2)
##
## Call:
## lm(formula = Total.Looking.Time ~ Age.Group * Face.Race * Music.Emotion,
## data = BabyData)
##
## Residuals:
## Min 1Q Median 3Q Max
## -24.8431 -8.0316 -0.1786 8.2809 27.7472
##
## Coefficients:
## Estimate Std. Error t value
## (Intercept) 28.0472 4.5406 6.177
## Age.Group -0.5167 0.7144 -0.723
## Face.RaceChinese -11.5424 6.6960 -1.724
## Music.Emotionsad -15.2955 6.6960 -2.284
## Age.Group:Face.RaceChinese 3.0376 1.0229 2.970
## Age.Group:Music.Emotionsad 3.4057 1.0229 3.330
## Face.RaceChinese:Music.Emotionsad 26.8342 9.3820 2.860
## Age.Group:Face.RaceChinese:Music.Emotionsad -5.7421 1.4252 -4.029
## Pr(>|t|)
## (Intercept) 4.16e-09 ***
## Age.Group 0.47045
## Face.RaceChinese 0.08645 .
## Music.Emotionsad 0.02351 *
## Age.Group:Face.RaceChinese 0.00338 **
## Age.Group:Music.Emotionsad 0.00105 **
## Face.RaceChinese:Music.Emotionsad 0.00473 **
## Age.Group:Face.RaceChinese:Music.Emotionsad 8.22e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 11.72 on 182 degrees of freedom
## Multiple R-squared: 0.1742, Adjusted R-squared: 0.1425
## F-statistic: 5.486 on 7 and 182 DF, p-value: 9.76e-06
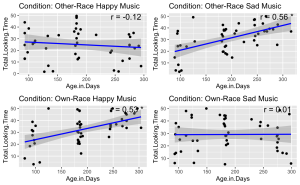
9. Given the significant three-way interaction, conduct Pearson correlation analyses to examine the linear relationship between total face looking time and participant age in days in each condition.
unique_conditions <- unique(BabyData$Condition) #Get unique conditions
correlation_results <- list() ## Initialize a list to store results
# Loop through each condition and perform Pearson correlation
for (condition in unique_conditions) {
# Subset data for the current condition
subset_data <- subset(BabyData, Condition == condition)
subset_data$Age.Group <- as.numeric(as.character(subset_data$Age.Group))
# Perform Pearson correlation
correlation_test <- cor.test(subset_data$Age.Group, subset_data$Total.Looking.Time, method = "pearson")
# Store the result
correlation_results[[condition]] <- correlation_test
}
# Print the results
correlation_results
## $`Other-Race Happy Music`
##
## Pearson's product-moment correlation
##
## data: subset_data$Age.Group and subset_data$Total.Looking.Time
## t = -0.64059, df = 43, p-value = 0.5252
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.3799180 0.2020743
## sample estimates:
## cor
## -0.09722666
##
##
## $`Other-Race Sad Music`
##
## Pearson's product-moment correlation
##
## data: subset_data$Age.Group and subset_data$Total.Looking.Time
## t = 4.4535, df = 46, p-value = 5.356e-05
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.3136678 0.7206311
## sample estimates:
## cor
## 0.5488839
##
##
## $`Own-Race Happy Music`
##
## Pearson's product-moment correlation
##
## data: subset_data$Age.Group and subset_data$Total.Looking.Time
## t = 3.8943, df = 46, p-value = 0.0003166
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## 0.2490419 0.6851408
## sample estimates:
## cor
## 0.4979416
##
##
## $`Own-Race Sad Music`
##
## Pearson's product-moment correlation
##
## data: subset_data$Age.Group and subset_data$Total.Looking.Time
## t = 0.25438, df = 47, p-value = 0.8003
## alternative hypothesis: true correlation is not equal to 0
## 95 percent confidence interval:
## -0.2466891 0.3149919
## sample estimates:
## cor
## 0.03707966
10. Visualize the relationship between total face looking time and participant age in days, categorized by different experimental conditions. Each condition should be represented in its own panel within a single figure. Additionally, for each panel:
# Get unique conditions
conditions <- unique(BabyData$Condition)
# Create a list to store plots
plot_list <- list()
# Loop through each condition and create a plot
for (condition in conditions) {
# Subset data for the condition
subset_data <- subset(BabyData, Condition == condition)
# Perform linear regression
fit <- lm(Total.Looking.Time ~ Age.in.Days, data = subset_data)
# Calculate Pearson correlation
cor_test <- cor.test(subset_data$Age.in.Days, subset_data$Total.Looking.Time)
# Create a scatter plot with regression line
p <- ggplot(subset_data, aes(x = Age.in.Days, y = Total.Looking.Time)) +
geom_point() +
geom_smooth(method = 'lm', color = 'blue') +
ggtitle(paste('Condition:', condition)) +
annotate("text", x = Inf, y = Inf, label = paste('r =', round(cor_test$estimate, 2), ifelse(cor_test$p.value < 0.05, "*", "")),
hjust = 1.1, vjust = 1.1, size = 5)
# Add plot to list
plot_list[[condition]] <- p
}
do.call(grid.arrange, c(plot_list, ncol = 2))
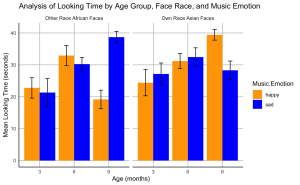
11. Analyze the impact of music emotional valence on the looking time for own- and other-race faces among different infant age groups (3, 6, and 9 months). Specifically, you are required to perform a series of independent sample t-tests.
# Ensure Age.Group is treated as a factor
BabyData$Age.Group <- as.factor(BabyData$Age.Group)
# Perform t-tests for each combination of Age.Group, Music.Emotion, and Face.Race
results <- list()
for(age_group in levels(BabyData$Age.Group)) {
for(music_emotion in unique(BabyData$Music.Emotion)) {
# Filter data for specific age group and music emotion
subset_data <- BabyData %>%
filter(Age.Group == age_group, Music.Emotion == music_emotion)
# Perform the t-test comparing Total.Looking.Time for own- vs. other-race faces
t_test_result <- t.test(Total.Looking.Time ~ Face.Race, data = subset_data)
# Store the results
result_name <- paste(age_group, music_emotion, sep="_")
results[[result_name]] <- t_test_result
}
}
# Print results
print(results)
## $`3_happy`
##
## Welch Two Sample t-test
##
## data: Total.Looking.Time by Face.Race
## t = -0.3153, df = 22.294, p-value = 0.7555
## alternative hypothesis: true difference in means between group African and group Chinese is not equal to 0
## 95 percent confidence interval:
## -12.465591 9.173257
## sample estimates:
## mean in group African mean in group Chinese
## 22.76875 24.41492
##
##
## $`3_sad`
##
## Welch Two Sample t-test
##
## data: Total.Looking.Time by Face.Race
## t = -1.0492, df = 22.86, p-value = 0.3051
## alternative hypothesis: true difference in means between group African and group Chinese is not equal to 0
## 95 percent confidence interval:
## -17.297369 5.658457
## sample estimates:
## mean in group African mean in group Chinese
## 21.32725 27.14671
##
##
## $`6_happy`
##
## Welch Two Sample t-test
##
## data: Total.Looking.Time by Face.Race
## t = 0.43226, df = 27.324, p-value = 0.6689
## alternative hypothesis: true difference in means between group African and group Chinese is not equal to 0
## 95 percent confidence interval:
## -6.401791 9.821475
## sample estimates:
## mean in group African mean in group Chinese
## 32.90100 31.19116
##
##
## $`6_sad`
##
## Welch Two Sample t-test
##
## data: Total.Looking.Time by Face.Race
## t = -0.62019, df = 27.075, p-value = 0.5403
## alternative hypothesis: true difference in means between group African and group Chinese is not equal to 0
## 95 percent confidence interval:
## -9.635393 5.162123
## sample estimates:
## mean in group African mean in group Chinese
## 30.20163 32.43827
##
##
## $`9_happy`
##
## Welch Two Sample t-test
##
## data: Total.Looking.Time by Face.Race
## t = -6.0414, df = 21.29, p-value = 5.08e-06
## alternative hypothesis: true difference in means between group African and group Chinese is not equal to 0
## 95 percent confidence interval:
## -27.28467 -13.31931
## sample estimates:
## mean in group African mean in group Chinese
## 19.13607 39.43806
##
##
## $`9_sad`
##
## Welch Two Sample t-test
##
## data: Total.Looking.Time by Face.Race
## t = 3.0179, df = 26.642, p-value = 0.005546
## alternative hypothesis: true difference in means between group African and group Chinese is not equal to 0
## 95 percent confidence interval:
## 3.335708 17.533234
## sample estimates:
## mean in group African mean in group Chinese
## 38.68853 28.25406
12. Create a bar plot to visualize the effects of music emotional valence on the looking time of infants at different ages for own- and other-race faces.
# Calculate means and standard errors
data_summary <- BabyData %>%
group_by(Age.Group, Face.Race, Music.Emotion) %>%
summarize(Mean = mean(Total.Looking.Time),
SE = sd(Total.Looking.Time)/sqrt(n())) %>%
ungroup()
## `summarise()` has grouped output by 'Age.Group', 'Face.Race'. You can override
## using the `.groups` argument.
# Create the bar plot
ggplot(data_summary, aes(x = factor(Age.Group), y = Mean, fill = Music.Emotion)) +
geom_bar(stat = "identity", position = position_dodge()) +
geom_errorbar(aes(ymin = Mean - SE, ymax = Mean + SE),
position = position_dodge(0.9), width = 0.25) +
scale_fill_manual(values = c("happy" = "orange", "sad" = "blue")) +
facet_wrap(~ Face.Race, scales = "free_x", labeller = labeller(Face.Race = c(Chinese = "Own Race Asian Faces", African = "Other Race African Faces"))) +
labs(x = "Age (months)", y = "Mean Looking Time (seconds)", title = "Analysis of Looking Time by Age Group, Face Race, and Music Emotion") +
theme_minimal() +
theme(
panel.grid.minor = element_blank(),
panel.grid.major = element_line(color = "gray", size = 0.5, linetype = "solid"), # Major grid lines
axis.line = element_line(color = "black", size = 0.5) # Axis lines
)
Welcome to the Perception and Sensorimotor Lab at McMaster University. As a budding cognitive psychologist here, you are about to embark on an explorative journey into the depth effect—a captivating psychological phenomenon that suggests visual events occurring in closer proximity (near space) are processed more efficiently than those farther away (far space). This effect provides a unique window into the cognitive architecture underpinning our sensory experiences, possibly implicating the involvement of the dorsal visual stream, which processes spatial relationships and movements in near space, and the ventral stream, known for its role in recognizing detailed visual information.
Your goal is to dissect whether the depth effect is task-dependent, aligning strictly with the dorsal/ventral stream dichotomy, or whether it represents a universal processing advantage for stimuli in near space across various cognitive tasks.
Your research journey begins in your lab. Imagine the lab as a gateway to a three-dimensional world, where the concept of depth is not only a subject of study but also a lived sensory experience for your participants! Seated inside a darkened tent, each participant grips a steering wheel, their primary tool for interaction and inputting responses. Before them, a screen comes to life with a 3D virtual environment meticulously engineered to test the frontiers of depth perception.
The virtual landscape participants encounter is a model of simplicity and complexity; as illustrated in the figure below, before the participants a ground plane extends into the depth of the screen, intersected by two sets of upright placeholder walls at varying depths—near and far. The walls stand on either side of the central axis, mirrored perfectly across the midline. The textures of the ground and placeholders—a random dot matrix and a checkerboard pattern, respectively—maintain a consistent density. These visual hints, alongside the textural gradients and the retinal size variance between near and far objects, act as subtle cues for depth perception.

From their first-person point of view, participants are asked to:
Through this experiment, you are not just observing the depth effect; you are dissecting it, unearthing the cognitive processes that allow humans to navigate the intricate dance of depth in our daily lives!
Let’s begin by loading the required libraries and the dataset. To do so download the file “NearFarRep_Outlier.csv” and run the following code.
# Loading the required
libraries library(tidyverse) # for data manipulation
library(rstatix) # for statistical analyses
library(emmeans) # for pairwise comparisons
library(afex) # for running anova using aov_ez and aov_car
library(kableExtra) # formatting html ANOVA tables
library(ggpubr) # for making plots
library(grid) # for plots
library(gridExtra) # for arranging multiple ggplots for extraction
library(lsmeans) # for pairwise comparisons
Read in the downloaded dataset “NearFarRep_Outlier.csv” as “NearFarData”. Remember to replace ‘path_to_your_downloaded_file’ with the actual path to the dataset on your system.
NearFarData <- read.csv('path_to_your_downloaded_file/NearFarRep_Outlier.csv')
The dataset contains the response times of participants and includes the following columns:
Please complete the accompanying exercises to the best of your abilities.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=34#h5p-4
1. Display the first few rows to understand your dataset. Display all column names in the dataset.
head(NearFarData) #Displaying the first few rows## X ID Response Con TarRT
## 1 1 10 Loc Near 0.6200754
## 2 2 10 Loc Near 0.2219719
## 3 3 1 Loc Near 0.2270377
## 4 4 9 Loc Near 0.5270686
## 5 5 25 Loc Near 0.2272455
## 6 6 18 Loc Near 0.2292785
colnames(NearFarData)
## [1] "X" "ID" "Response" "Con" "TarRT"
2. Set up “Response” and “Con” as factors, then check the structure of your data to make sure your factors and levels are set up correctly.
NearFarData <- NearFarData %>% convert_as_factor(Response, Con)str(NearFarData)## 'data.frame': 11154 obs. of 5 variables:
## $ X : int 1 2 3 4 5 6 7 8 9 10 ...
## $ ID : int 10 10 1 9 25 18 4 9 8 18 ...
## $ Response: Factor w/ 2 levels "Disc","Loc": 2 2 2 2 2 2 2 2 2 2 ...
## $ Con : Factor w/ 2 levels "Far","Near": 2 2 2 2 2 2 2 2 2 2 ...
## $ TarRT : num 0.62 0.222 0.227 0.527 0.227 ...
3. Perform basic data checks for missing values and data consistency.
sum(is.na(NearFarData)) # Checking for missing values in the dataset## [1] 0
4. Convert the values in your dependent measures column “TarRT” to seconds.
NearFarData$TarRT <- NearFarData$TarRT * 1000
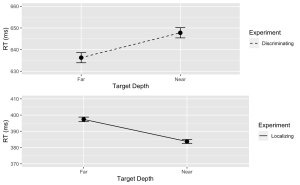
# Calculate means and standard errors for each combination of 'Response' and 'Con'summary_df <- NearFarData %>% group_by(Response, Con) %>% summarise( MeanRT = mean(TarRT), SERT = sd(TarRT) / sqrt(n()) )6. Using the “ggplot2” package, create a line plot with error bars for the Discrimination task.
# Now, using ggplot to create the plotDisc.plot <- ggplot(data = filter(summary_df, Response=="Disc"), aes(x = Con, y = MeanRT, group = Response)) + geom_line(aes(linetype = "Discriminating")) + # Add a linetype aesthetic geom_errorbar(aes(ymin = MeanRT - SERT, ymax = MeanRT + SERT), width = 0.1) + geom_point(size = 3) + theme_gray() + labs( x = "Target Depth", y = "RT (ms)", color = "Experiment", linetype = "Experiment") + scale_linetype_manual(values = "dashed") + # Set the linetype for "Disc" to dashed ylim(630, 660) # Set the y-axis limits
7. Similarly, create a line plot with error bars for the Localization task. Use a dashed line for this plot with the following exceptions:
Loc.plot <- ggplot(data = filter(summary_df, Response=="Loc"), aes(x = Con, y = MeanRT, group = Response)) +
geom_line(aes(linetype = "Localizing")) + # Add a linetype aesthetic
geom_errorbar(aes(ymin = MeanRT - SERT, ymax = MeanRT + SERT), width = 0.1) +
geom_point(size = 3) +
theme_gray() +
labs(
x = "Target Depth",
y = "RT (ms)",
color = "Experiment",
linetype = "Experiment") +
scale_linetype_manual(values = "solid") + # Set the line type for "Disc" to dashed
ylim(370, 410) # Set the y-axis limits
8. Finally, use the grid.arrange() function from the “gridExtra” package to stack the plots for the Discrimination and Localization tasks on top of each other.
grid.arrange(Disc.plot, Loc.plot, ncol = 1) # Stack the plots on top of each other
9. Using the “anova_test” function, conduct a two-way between-subjects ANOVA to investigate the effects of Con (Condition) and Response (Task type) on the target response times (TarRT). After running the ANOVA, use the “get_anova_table” function to present the results.
anova <- anova_test(
data = NearFarData, dv = TarRT, wid = ID,
between = c(Con, Response), detailed = TRUE, effect.size = "pes")
## Warning: The 'wid' column contains duplicate ids across between-subjects
## variables. Automatic unique id will be created
get_anova_table(anova)
## ANOVA Table (type III tests)
##
## Effect SSn SSd DFn DFd F p p<.05
## 1 (Intercept) 2.972143e+09 111582983 1 11150 296993.268 0.00e+00 *
## 2 Con 3.185839e+03 111582983 1 11150 0.318 5.73e-01
## 3 Response 1.762985e+08 111582983 1 11150 17616.741 0.00e+00 *
## 4 Con:Response 4.390483e+05 111582983 1 11150 43.872 3.67e-11 *
## pes
## 1 9.64e-01
## 2 2.86e-05
## 3 6.12e-01
## 4 4.00e-03
## Fitting a linear model to datalm_model <- lm(TarRT ~ Con * Response, data = NearFarData)
# Get the estimated marginal means
emm <- emmeans(lm_model, specs = pairwise ~ Con * Response)
# View the results
print(post_hoc_results)
## contrast estimate SE df t.ratio p.value
## Far Disc - Near Disc -11.5 2.70 11150 -4.250 0.0001
## Far Disc - Far Loc 238.9 2.67 11150 89.348 <.0001
## Far Disc - Near Loc 252.6 2.67 11150 94.581 <.0001
## Near Disc - Far Loc 250.4 2.69 11150 93.131 <.0001
## Near Disc - Near Loc 264.0 2.68 11150 98.341 <.0001
## Far Loc - Near Loc 13.6 2.66 11150 5.124 <.0001
##
## P value adjustment: tukey method for comparing a family of 4 estimates
You are a researcher in the EdCog Lab at McMaster University. The Lab is conducting a study aimed at understanding the beliefs of instructors about student abilities in STEM (Science, Technology, Engineering, and Math) disciplines. This study is motivated by a growing body of literature suggesting that instructors’ beliefs about intelligence and success—categorized into brilliance belief (the idea that success requires innate talent), universality belief (the belief that success is achievable by everyone versus only a select few), and mindset beliefs (the view that intelligence and skills are either fixed or can change over time)—play a crucial role in educational practices and student outcomes. Understanding these beliefs is particularly important in STEM fields, where perceptions of innate talent versus learned skills can significantly influence teaching approaches and student engagement.
The survey was distributed through LimeSurvey to instructors across the Science, Health Sciences, and Engineering faculties. Participants were asked a series of Likert-scale questions (ranging from strongly disagree to strongly agree) aimed at assessing their beliefs in each of the three areas. Additional demographic and background questions were included to control for variables such as years of teaching experience, field of specialization, and level of education.
The sample data file (“EdCogData.xlsx) for this exercise is structured as such:
Let’s begin by running the following code in RStudio to load the required libraries. Make sure to read through the comments embedded throughout the code to understand what each line of code is doing.
# Here we create a list called "my_packages" with all of our required libraries
my_packages <- c("tidyverse", "readxl", "xlsx", "dplyr", "ggplot2")
# Checking and extracting packages that are not already installed
not_installed <- my_packages[!(my_packages %in% installed.packages()[ , "Package"])]
# Install packages that are not already installed
if(length(not_installed)) install.packages(not_installed)
# Loading the required libraries
library(tidyverse) # for data manipulation
library(dplyr) # for data manipulation
library(readxl) # to read excel files
library(xlsx) # to create excel files
library(ggplot2) # for making plots
Make sure to have the required dataset (“EdCogData.xlsx“) for this exercise downloaded. Set the working directory of your current R session to the folder with the downloaded dataset. You may do this manually in R studio by clicking on the “Session” tab at the top of the screen, and then clicking on “Set Working Directory”.
If the downloaded dataset file and your R session are within the same file, you may choose the option of setting your working directory to the “source file location” (the location where your current R session is saved). If they are in different folders then click on “choose directory” option and browse for the location of the downloaded dataset.
You may also do this by running the following code:
setwd(file.choose())
Once you have set your working directory either manually or by code, in the Console below you will see the full directory of your folder as the output.
Read in the downloaded dataset as “edcogData” and complete the accompanying exercises to the best of your abilities.
# Read xlsx file
edcog = read_excel("EdCogData.xlsx")
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=39#h5p-5
Note: Shaded boxes hold the R code, while the white boxes display the code’s output, just as it appears in RStudio. The “#” sign indicates a comment that won’t execute in RStudio.
Load the dataset into RStudio and inspect its structure.
head(edcogData) # View the first few rows of the datasetncol(edcogData) #Q1#[1] 26
colnames(edcogData) #Q2
#[1] "ID" "Brilliance1" "Brilliance2" "Brilliance3" "Brilliance4"
#[6] "Brilliance5" "MindsetGrowth1" "MindsetGrowth2" "MindsetGrowth3" "MindsetGrowth4"
#[11] "MindsetGrowth5" "MindsetFixed1" "MindsetFixed2" "MindsetFixed3" "MindsetFixed4"
#[16] "MindsetFixed5" "Nonuniversality1" "Nonuniversality2" "Nonuniversality3" "Nonuniversality4"
#[21] "Nonuniversality5" "Universality1" "Universality2" "Universality3" "Universality4"
#[26] "Universality5"
Prepare the data for analysis by ensuring it is in the correct format.
1. Are there any missing values in the dataset?
sum(is.na(edcogData))
[1] 0
1. Create aggregate scores for each dimension (Brilliance, Fixed, Growth, Nonuniversal, Universal).
edcogData$Brilliance <- rowMeans(edcogData[,c("Brilliance1", "Brilliance2", "Brilliance3", "Brilliance4", "Brilliance5")])
edcogData$Growth <- rowMeans(edcogData[,c("MindsetGrowth1", "MindsetGrowth2", "MindsetGrowth3", "MindsetGrowth4", "MindsetGrowth5")])
edcogData$Fixed <- rowMeans(edcogData[,c("MindsetFixed1", "MindsetFixed2", "MindsetFixed3", "MindsetFixed4", "MindsetFixed5")])
edcogData$Universal <- rowMeans(edcogData[,c("Universality1", "Universality2", "Universality3", "Universality4", "Universality5")])
edcogData$Nonuniversal <- rowMeans(edcogData[,c("Nonuniversality1", "Nonuniversality2", "Nonuniversality3", "Nonuniversality4", "Nonuniversality5")])
2. Create a new data frame named “edcog.agg.wide” that contains only the ID column and the aggregated score columns from “edcogData”.
edcog.agg.wide <- edcogData %>% select(ID, Brilliance, Fixed, Growth, Nonuniversal, Universal)
3. Convert “edcog.agg.wide” from a wide to a long format named “edcog.agg.long”, with the following columns:
edcog.agg.long <- edcog.agg.wide %>%
select(ID, Brilliance, Fixed, Growth, Nonuniversal, Universal) %>%
pivot_longer(
cols = -ID, # Select all columns except for ID
names_to = "Dimension",
values_to = "AggregateScore" )
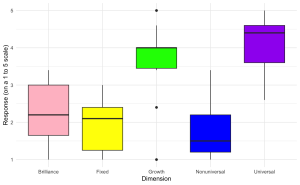
1. Create a boxplot to visualize the distribution of aggregate scores across different dimensions (Brilliance, Fixed, Growth, Nonuniversal, Universal) from the survey data with the following specifications:
ggplot(edcog.agg.long, aes(x = Dimension, y = AggregateScore, fill = Dimension)) +
geom_boxplot() +
scale_fill_manual(values = c("Brilliance" = "pink", "Fixed" = "yellow",
"Growth" = "green", "Nonuniversal" = "blue",
"Universal" = "purple")) +
labs(y = "Response (on a 1 to 5 scale)", x = "Dimension") +
theme_minimal() +
theme(legend.position = "none") # Hide the legend since color coding is evident
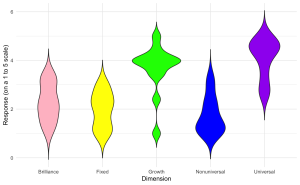
2. Generate a violin plot to visualize the distribution of aggregate scores for different dimensions (Brilliance, Fixed, Growth, Nonuniversal, Universal) from the survey data with the following specifications:
ggplot(edcog.agg.long, aes(x = Dimension, y = AggregateScore, fill = Dimension)) +
geom_violin(trim = FALSE) +
scale_fill_manual(values = c("Brilliance" = "pink", "Fixed" = "yellow",
"Growth" = "green", "Nonuniversal" = "blue",
"Universal" = "purple")) +
labs(y = "Response (on a 1 to 5 scale)", x = "Dimension") +
theme_minimal() + theme(legend.position = "none")
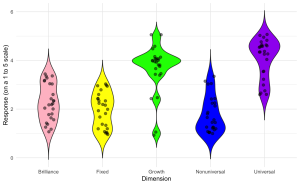
3. Enhance the violin plot by overlaying individual data points to show the raw data distribution alongside the aggregated density estimates.
ggplot(edcog.agg.long, aes(x = Dimension, y = AggregateScore, fill = Dimension)) +
geom_violin(trim = FALSE) +
geom_jitter(width = 0.1, size = 2, alpha = 0.5) + # Adjust 'width' for jittering, 'size' for point size, and 'alpha' for transparency
scale_fill_manual(values = c("Brilliance" = "pink", "Fixed" = "yellow",
"Growth" = "green", "Nonuniversal" = "blue",
"Universal" = "purple")) +
labs(y = "Response (on a 1 to 5 scale)", x = "Dimension") +
theme_minimal() +
theme(legend.position = "none")
You are a clinical researcher at the Hamilton General Hospital, and your lab is studying how people grieve and cope with the loss of a loved one. Specifically, some people ruminate when they grieve, and so your lab is interested in understanding the different ways in which this grief-related rumination manifests in people. Andrews et al. (2021) recently developed the Bereavement Analytical Rumination Questionnaire (BARQ) to evaluate two dimensions of rumination: 1) the cause of the loss (i.e., root cause analysis – RCA) and 2) how an individual reinvests their time meaningfully following the loss (i.e., reinvestment analysis – RIA).
Your lab is curious about the following questions:
Similar to Andrews et al. (2021), your lab decides to collect the following information from a questionnaire distributed to 50 respondents:
To answer the lab’s questions, please run the following analyses.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-6
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-54
2. Conduct a confirmatory factor analysis (CFA) on the seven items of the BARQ. Items 1-4 should form the latent factor RCA, and items 5-7 should form the latent factor RIA.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-55
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-56
3. Compare RCA and RIA latent factor means between the following groups. Which comparisons have statistically significant differences in the RCA and RIA means?
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-57
4. The three time variables in the questionnaire may exhibit multilinearity with one another: age of the deceased, current age of the respondent, and the time passed since death. For instance, the age of the deceased and the age of the respondent may be collinear with one another, especially if the relationship of the deceased to respondent is that of a child. To test for multilinearity, assess the variance inflation factor (VIF) of a regression model that includes the three time variables as predictors for RCA and RIA. A VIF of 1 means there is no correlation between predictor variables, while a VIF above 5 indicates a high correlation.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-58
5. Create a scatterplot of the participant’s latent RCA factor score (y-axis) against the age of the deceased child (x-axis).
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=41#h5p-59
Andrews, P. W., Altman, M., Sevcikova, M., & Cacciatore, J. (2021). An evolutionary approach to grief-related rumination: Construction and validation of the Bereavement Analytical Rumination Questionnaire. Evolution and Human Behavior, 42(5), 441-452.
Hollywood studios and movie producers are keenly interested in determining what types of story scripts will resonate with audiences and critics. A few budding screenwriters approach you, a social psychology researcher specializing in qualitative content analysis, to conduct an analysis of movie plots in order to help them determine the types of storylines that may appeal to mainstream viewers. 150 movies from the last five years were randomly selected and analyzed for plot and character traits. You decide to analyze the following six narrative characteristics for the exploratory study: 1) genre, 2) plot shape, 3) protagonist goal type, 4) protagonist agency, 5) protagonist cooperativeness, and 6) protagonist assertiveness. You are interested to see how these characteristics relate to the following outcomes: 1) average critic rating of the movie (as a percentage score) and 2) the net profit of the movie (in US dollars).
To analyze these six narrative characteristics, you adopt Brown & Tu’s (2020) scheme for plot and Berry & Brown’s (2017) classification scheme for literary characters. The coding scheme for the five narrative characteristics are as follows:
| Label | Code |
| Drama | 1 |
| Comedy | 2 |
| Romance | 3 |
| Action | 4 |
| Horror | 5 |
| Label | Code |
| Fall-Rise | 1 |
| Fall-Rise-Fall | 2 |
| Rise-Fall | 3 |
| Rise-Fall-Rise | 4 |
| Label | Code |
| Striving | 1 |
| Coping | 2 |
| Label | Code |
| High | 1 |
| Medium | 2 |
| Low | 3 |
| Label | Code |
| High | 1 |
| Medium | 2 |
| Low | 3 |
You also recruit a second coder in order to determine whether there is inter-rater reliability in your coding method.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-60
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-61
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-62
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-63
4. Visualize these plot shape distributions across each genre in a grouped bar plot. Be sure to label the y-axis, x-axis, and legend.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-64
5. Run comparative analyses to see which, if any, of the six characteristics are related to one another. Because the six characteristics are not normally distributed, use non-parametric tests, such as Chi-Square. For example, is the relationship between genre and plot statistically significant?
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-65
6. Run an analysis of variance test to see which, if any, of the five rater-coded variables are related with either of the two outcome variables. Consider if you should run a 2-way or 3-way factorial ANOVA. What assumptions do you need to consider and test before running an ANOVA?
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-66
7. Run a cluster analysis to see if which, if any, of the categories in the six narrative characteristics variables cluster together.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-67
8. Determine the inter-rater reliability between the two coders for each of the five rater-coded variables. A Cohen’s Kappa score of greater than 0.8 reflects strong inter-rate agreement.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=43#h5p-68
Berry, M., & Brown, S. (2017). A classification scheme for literary characters. Psychological Thought, 10(2).
Brown, S., & Tu, C. (2020). The shapes of stories: A “resonator” model of plot structure. Frontiers of Narrative Studies, 6(2), 259-288.
A new science fiction massive multiplayer online role-playing game (MMORPG) was released that allows players to design the sound of their avatar’s voice. The game features a sliding pitch scale for players to select the fundamental pitch (f0) and formant frequencies (pf) (both measured in hertz) of their avatar’s voice. As players engage with the game world and interact with other players through their avatar, specific personality traits for their avatar would emerge. In order to determine whether the avatars created within this science fiction game world resemble the vocal and personality patterns found in the real-world, the designers of the game collaborated with psychologists to investigate this comparison. The game designers were particularly inspired by Stern et al. (2021)’s study that explored how voice pitch is related to self-reported extraversion, dominance, and sociosexuality in men and women.
The current video game study recruited a sample of 475 players from the game, 200 men and 275 women, all aged from 20 to 45. In Stern et al. (2021)’s study, participants would read short text passages so that the researchers could determine the fundamental pitch and formant frequencies of the participants’ voice. However, the game designers are able to extract the information about an avatar’s voice from the selections that the player made on the sliding pitch scale when the player created their avatar. The game designers asked the 475 players to each fill out a questionnaire about their avatar’s personality traits (Likert ratings on a scale from 1-5 from the Big 5), dominance (Likert ratings on a scale from 1-5 from the Interpersonal Adjective List, and sociosexuality (Likert ratings on a scale from 1-5 from the SOI-R). The sociosexuality score is the mean of the ratings for the three dimensions of sociosexuality: behaviour, attitude, and desire. The game designers have already calculated the Cronbach’s alpha for the raw scores of the scales and found that there was good reliability. As certain MMORPGS can provide the potential for players to explore and express their sexuality through their avatars, such as the current new science fiction MMORPG that is the focus of this scenario, the game designers are curious as to whether there is a relationship between these self-reported traits and the type of voice that the player has designed for their avatar.
To help the game designers investigate this, please answer the following questions:
Load the data. Data can be found in voiceData.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=45#h5p-69
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=45#h5p-70
2. Create violin plots comparing gender with f0, pf, and the nine personality trait variables. Visually inspect the violin plots and answer the following questions:
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=45#h5p-71
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=45#h5p-72
Stern, J., Schild, C., Jones, B. C., DeBruine, L. M., Hahn, A., Puts, D. A., … & Arslan, R. C. (2021). Do voices carry valid information about a speaker’s personality?. Journal of Research in Personality, 92, 104092.
You are a researcher at an academy of music in Ontario. You would like to understand how musicians in a string quartet coordinate and synchronize with one another. To do this, you recruit two groups of musicians to the Large Interactive Virtual Environment laboratory (LIVELab, https://livelab.mcmaster.ca) at McMaster University where the musician’s live behavioural data can be recorded. Specifically, body sway was measured using an infrared optical motion capture system, which requires each musician to wear a felt cap with reflective markers.
The two groups of musicians are two string quartets. A string quartet consists of a first violinist (labeled as M1), a second violinist (M2), one violist (M3), and one cellist (M4). Both string quartets performed the same 2-minute musical excerpt three times (i.e., three trials). One string quartet performed the excerpt in the same room (the “sight” condition), while the other string quartet performed in a room where there were dividers between the musicians to prevent them from seeing one another (the “no sight” condition).
Body sway was measured as the change of position in millimeters in the anterior-posterior direction. The data can be found below. Measurements were taken at a frequency of 8hz (i.e., 8 time samples per second). A 2-minute recording will therefore yield 960 time sample data points per musician.
Musical synchrony and interpersonal coordination can be analyzed by comparing how similar a musician’s body sway time series is to another musician’s time series (i.e., a cross-correlation between two time series) as well as whether one musician’s time series is able to predict another musician’s time series (i.e., granger causality analysis between time series), which is also known as the information flow.
As a researcher of the academy of music, you would like to understand the following questions:
To answer the above question, please complete the following statistical analyses:
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=47#h5p-73
2. Convert the timeline data of each musician to z-scores.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=47#h5p-74
3. Run a window cross-correlation analysis between the following pairs of musicians in each trial and in each condition (there are six possible pair combinations):
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=47#h5p-76
4. Run a granger causality analysis between the following pairs of musicians in each trial and each condition (there are 12 possible pair combinations)
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=47#h5p-77
Wood, E. A., Chang, A., Bosnyak, D., Klein, L., Baraku, E., Dotov, D., & Trainor, L. J. (2022). Creating a shared musical interpretation: Changes in coordination dynamics while learning unfamiliar music together. Annals of the New York Academy of Sciences, 1516(1), 106-113.
You are a social perceptions researcher and are interested in whether there are differences in how children from Country A and children from Country B perceive nationality. You were specifically inspired by Siddiqui, Cimpian, and Rutherford (2020)’s study comparing the degree of essentialist perspectives of national groups between Canadian and American children.
To investigate whether there are any differences in essentialist views about national groups, you recruited 50 children from the ages of 5-8 years old from Country A and 50 children from the ages of 4-9 years old from Country B.
You ask the children to answer questions pertaining to the following categories of essentialism:
The response options were binary for each question, where one answer would receive a score of 1, while another answer would receive a score of zero. Questions within the same category were averaged to give an essentialism score for that category. Scores that are closer to 1 indicate high essentialism. The responses from the children can be found below.
To compare whether essentialist views differ between children from Country A and Country B, please run the following statistical analyses:
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=49#h5p-78
2. Create a scatterplot graph to show the essentialism scores
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=49#h5p-79
3. Run pairwise t-test comparisons between the essentialism scores for each of the seven categories between Country A and Country B.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=49#h5p-80
4. Because there are multiple comparisons, the risk of type-1 error is increased. Run a Bonferroni correction to account for this.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=49#h5p-81
5. Create a mixed effects linear model using the categories of essentialism as a categorical predictor and Children’s age as a continuous predictor.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=49#h5p-82
Siddiqui, H., Cimpian, A., & Rutherford, M. D. (2020). Canadian children’s concepts of national groups: A comparison with children from the United States. Developmental psychology, 56(11), 2102.
II
Electroencephalography, or EEG, is a non-invasive neuroimaging technique that measures and records the electrical activity of the brain. It involves placing electrodes on the scalp to detect the voltage fluctuations resulting from ionic current within the neurons of the brain. EEG provides a real-time representation of brain activity and is widely used in computational neuroscience to study neural processes and understand brain function.
In computational neuroscience, EEG data is analyzed using advanced algorithms and mathematical models to extract valuable information about cognitive processes, sensory perception, and various neurological disorders. Researchers use EEG to investigate patterns of neural activity, brain connectivity, and the temporal dynamics of information processing. The versatility and temporal precision of EEG make it a valuable tool for studying brain function and contributing to our understanding of the complex interplay of neurons in the human brain.
EEG signals are characterized by different frequency bands that reflect the underlying neural activity. The frequency bands are defined based on the frequency range of the EEG signal, and each band is associated with a specific type of brain activity. The frequency bands are as follows:
Studying the interplay of these EEG bands provides valuable information about brain function, aiding researchers and clinicians in understanding cognitive processes, diagnosing disorders, and developing therapeutic interventions. EEG data analysis allows for the exploration of brain dynamics and can unlock new insights into the complexities of the human mind.
In EEG (Electroencephalography) data, channels refer to the specific locations on the scalp where electrodes are placed to record electrical activity produced by the brain. These electrodes are part of an EEG cap or array. Each channel corresponds to a unique electrode, and the signals collected from these channels collectively provide a comprehensive view of brain activity.
The placement of channels is crucial for capturing signals from different regions of the brain. Common international standards, such as the 10-20 system, are used to define the locations of these channels. The system is named after the fact that the distance between adjacent electrodes is approximately 20% of the total front-to-back or right-to-left distance of the skull, depending on the region. Each channel records the voltage fluctuations over time, reflecting the electrical activity of the neurons in the underlying brain region. Analyzing EEG data from multiple channels allows researchers and clinicians to examine the spatial distribution of brain activity and identify patterns associated with various cognitive states, disorders, or specific tasks.
In this section, we focus on analyzing EEG data using R. Even though EEG data analysis can be performed using both MATLAB and R, and the choice between the two often depends on the preferences of the researcher, the availability of specific toolboxes or packages, and the nature of the analysis, MATLAB is a more popular choice for EEG data analysis due to its wide range of packages for EEG data analysis.
Key R Packages:
There are various packages and tools to preprocess, visualize, and extract meaningful insights from the EEG recordings. Below is a brief overview of the some typical steps involved in EEG data analysis using R:
eegkit: A package for importing and preprocessing EEG data in R.eegUtils: A package for performing basic EEG preprocessing and plotting of EEG data.ERP: A package for analyzing, identifying and extracting event-related potentials (ERPs) related to specific stimuli or events in R.Key Software:
EEGLAB: A MATLAB toolbox for processing and analyzing EEG data. It includes a variety of functions for importing, preprocessing, visualizing, and analyzing EEG data. EEGLAB also provides a graphical user interface (GUI) for performing EEG data analysis.FieldTrip: A MATLAB toolbox for analyzing EEG and MEG data. It includes algorithms for simple and advanced analysis of MEG and EEG data, such as time-frequency analysis, source reconstruction using dipoles, distributed sources, and beamformers, connectivity analysis, and non-parametric statistical testing.Brainstorm: A MATLAB toolbox dedicated to the analysis of brain recordings: MEG, EEG, fNIRS, ECoG, depth electrodes and multiunit electrophysiologyThis process aims to remove noise, artifacts, and other unwanted elements while preserving the integrity of the neural signals. Effective preprocessing ensures reliable results in subsequent analyses, focusing on genuine neural activity. Here are key steps involved in EEG data preprocessing:
By implementing these preprocessing steps, researchers can enhance the quality of EEG data, reduce noise, and improve the accuracy of subsequent analyses, leading to more reliable insights into brain function and cognition.
Statistical analysis of EEG (Electroencephalography) data is crucial for drawing meaningful conclusions from experimental results. EEG experiments often involve comparing conditions, groups, or time points to uncover patterns of brain activity associated with specific cognitive processes or experimental manipulations. Here’s an overview of key considerations and methods in the statistical analysis of EEG data:
By employing these statistical approaches, researchers can draw robust conclusions from EEG data, uncover patterns, and elucidate the neurophysiological mechanisms underlying cognitive processes or clinical conditions.
# Install eegkit package
install.packages("eegkit")
# Load eegkit package
library(eegkit)# Load EEG data
data("eegdata")
# View the first 5 rows of the data
head(eegdata)eegsmooth Smooths single- or multi-channel electroencephalography (EEG) with respect to space and/or time
## get "PZ" electrode of "c" subjects
idx <- which(eegdata$channel=="PZ" & eegdata$group=="c")
eegdata1 <- eegdata[idx,]
## temporal smoothing eegmod <- eegsmooth(eegdata1$voltage,time=eegdata1$time) ## define data for prediction time <- seq(min(eegdata1$time),max(eegdata1$time),length.out=100) yhat <- predict(eegmod,newdata=time,se.fit=TRUE) ## plot results using eegtime eegtime(time*1000/255,yhat$fit,voltageSE=yhat$se.fit,ylim=c(-4,4),main="Pz")
 `eegtime` Creates plot of single-channel electroencephalography (EEG) time course with optional confidence
interval. User can control the plot orientation, line types, line colors, etc.
Example: Plotting a single channel
`eegtime` Creates plot of single-channel electroencephalography (EEG) time course with optional confidence
interval. User can control the plot orientation, line types, line colors, etc.
Example: Plotting a single channel
## get "PZ" electrode from "eegdata" data
idx <- which(eegdata$channel=="PZ")
eegdata2 <- eegdata[idx,]
## get average and standard error (note se=sd/sqrt(n))
eegmean <- tapply(eegdata2$voltage,list(eegdata2$time,eegdata2$group),mean)
eegse <- tapply(eegdata2$voltage,list(eegdata2$time,eegdata2$group),sd)/sqrt(50)
## plot results with legend
tseq <- seq(0,1000,length.out=256)
eegtime(tseq,eegmean[,2],voltageSE=eegse[,2],ylim=c(-10,6),main="Pz")
eegtime(tseq,eegmean[,1],vlty=2,vcol="red",voltageSE=eegse[,1],scol="pink",add=TRUE)
legend("bottomright",c("controls","alcoholics"),lty=c(1,2),
lwd=c(2,2),col=c("blue","red"),bty="n") Further Reading and References:
In a study examining EEG recordings, data from 10 alcoholics and 10 control subjects were collected during a 10-second experiment. The dataset comprises four columns: “ID,” “Group,” “Timestep,” and “Voltage.”
read.csv() command in R.Group is a factor with two levels: Control and Alcohol.Perform the following analyses using the loaded data:
This study aims to explore and statistically analyze the EEG data to determine if there are discernible differences in voltage levels between alcoholics and control subjects. The combination of exploratory visualization and statistical tests provides a comprehensive understanding of the EEG patterns and potential group distinctions.
# Read the CSV file
## Note: this is a simplified simulated EEG data from 10 alcoholic and 10 control subjects participating in a 10 second experiment
## the voltage is recorded with dt=0.01s
data <- read.csv("eeg.csv")
# Plot EEG of one participant
participant_id <- 1 # Change this to the desired participant ID
participant_data <- data[data$ID == participant_id, ]
# Plot EEG data
plot(participant_data$Timestep, participant_data$Voltage, type = "l",
main = paste("EEG Plot for Participant", participant_id),
xlab = "Time (s)", ylab = "Voltage")
# Calculate descriptive statistics for each group
control_group <- subset(data, Group == "Control")
alcoholic_group <- subset(data, Group == "Alcoholic")
# Calculate statistics for the control group
control_stats <- c(mean(control_group$Voltage), sd(control_group$Voltage),
min(control_group$Voltage), max(control_group$Voltage))
control_stats
# Calculate statistics for the alcoholic group
alcoholic_stats <- c(mean(alcoholic_group$Voltage), sd(alcoholic_group$Voltage),
min(alcoholic_group$Voltage), max(alcoholic_group$Voltage))
# Create a data frame to store group statistics
group_stats <- data.frame(Group = c("Control", "Alcoholic"),
Mean = c(control_stats[1], alcoholic_stats[1]),
SD = c(control_stats[2], alcoholic_stats[2]),
Min = c(control_stats[3], alcoholic_stats[3]),
Max = c(control_stats[4], alcoholic_stats[4]))
group_stats
# Perform a t-test between groups to determine if there is a statistically significant difference in the mean voltage values between the control group and the alcoholic group
t_test_result <- t.test(data$Voltage ~ data$Group)
t_test_result
# Perform ANOVA to compare
anova_result <- aov(Voltage ~ Group, data = data)
summary(anova_result)
Magnetic Resonance Imaging (MRI) is a pivotal medical imaging technique using nuclear magnetic resonance for producing detailed internal body structures, particularly effective for visualizing soft tissues with superior clarity compared to X-rays or CT scans. This modality is extensively applied in diagnosing a wide range of conditions, notably in brain imaging for detecting tumors, strokes, and other neurological conditions.
MRI involves a patient lying inside a large magnet, where radio waves target the body. The MRI sensors detect the energy emitted from the body and convert this data into images. Unlike methods involving ionizing radiation, MRI’s safety profile allows for repeated usage. However, its strong magnetic field may be contraindicated in patients with specific metal implants, and some may find the lengthy, motionless procedure challenging.
In the field of neuroscience, MRI’s non-invasive and detailed imaging capabilities are indispensable for accurately distinguishing brain tissues and detecting abnormalities, playing a critical role in diagnosing and monitoring neurological diseases like multiple sclerosis and Alzheimer’s.
In this section, we focus on visualizing and analyzing MRI data using R programming.
Data Format:
MRI images are commonly available in NIFTI format, with file extensions such as .nii or .nii.gz (compressed). NIFTI files are compatible with various neuroimaging analysis software.
Key R Packages:
oro.nifti: Essential for loading and manipulating NIFTI objects.neurobase: Extends oro.nifti capabilities, offering additional imaging functions.Loading MRI Data in R:
# Loading the oro.nifti and neurobase packages
library(oro.nifti)
library(neurobase)
# Reading a NIFTI file
mri_img = readnii("training01_01_mri_img.nii.gz")Visualizing MRI Data:
ortho2(mri_img)The neurobase::ortho2 function displays nifti objects in 3 different planes.
image(mri_img, useRaster= TRUE)This function from oro.nifti provides a lightbox view, showcasing all slices of an MRI image.
oro.nifti::slice(mri_img, z = c(60, 80))Viewing specific slices is vital for detailed examination of particular brain regions.
Analyzing Voxel Value Distributions:
In MRI imaging, voxels (short for “volumetric pixels”) function similarly to pixels in 2D images but they represent the smallest distinguishable three-dimensional units of the scanned volume.
Voxel values in MRI data can be analyzed to understand the distribution of different tissue types.
plot(density(mri_img))This plot helps in understanding the distribution of voxel intensities.
hist(mri_img)Histograms are useful for visualizing the frequency distribution of voxel intensities.
Segmenting Brain Regions (ROIs):
A critical aspect of MRI analysis in neuroscience is the segmentation of the brain into biologically significant regions of interest (ROIs). This includes tissue segmentation, identifying deep gray matter structures, and segmenting pathology such as multiple sclerosis lesions or tumors.
Further Reading and References:
For more detailed information and advanced techniques in MRI analysis using R, the following resources are recommended:
Preprocessing is a critical step in MRI image analysis, pivotal for ensuring accuracy and reliability of results. This process involves several key steps:
Preprocessing enhances image quality, interpretability, and increases the statistical power of analyses. It lays the groundwork for advanced analyses, including functional MRI studies and machine learning approaches.
While preprocessing is critical, it is not the focus of this tutorial. For detailed preprocessing methods of MRI images in R, taking a look at this tutorial from John Muschelli and Kristin Linn is highly recommended.
For the purposes of this tutorial, we will assume preprocessing is already completed.
Statistical analysis in MRI data encompasses a variety of techniques, each offering unique insights into brain structure and function:
Voxel-based morphometry is a popular technique in MRI analysis. This process involves focusing on specific regions of interest (ROIs) in the brain and analyzing the volume of these regions. The volume of a region can be calculated by multiplying the number of voxels in the region with the volume of each voxel.
While our primary focus isn’t on calculating the volume of a ROI, those interested can follow these steps to mask an ROI and calculate its volume. Note that these steps require specific software and libraries.
Ensure you have the necessary libraries installed. Installation of ANTsR is more complex than typical R packages. You can find detailed installation instructions here.
library(ANTsR)
library(oro.nifti)mri_image <- niftiImageRead("path_to_your_mri_image.nii", reorient = FALSE)segmentation_results <- atropos(a = mri_image, m = '[3,1x1x1]', c = '[2,0]', i = 'kmeans[3]', x = 1)csf <- segmentation_results$segmentation == 1
gm <- segmentation_results$segmentation == 2
wm <- segmentation_results$segmentation == 3
niftiImageWrite(csf, "csf_segmented.nii")
niftiImageWrite(gm, "gm_segmented.nii")
niftiImageWrite(wm, "wm_segmented.nii")vres = voxres(t1, units = "cm")
vol_csf = csf * vresConsider a study with 30 adults over 55 with Alzheimer’s and 30 controls. The study spans over 2 years, tracking hippocampal volume changes.
alzheimer_hippo_vol.csv using read.csv() command in R.condition is a factor with two levels: control and alzheimer.initial_vol column.loss column.Perform the following analyses using the loaded data:
1. Plotting Marginal Means of Diagnosis: Visualize the average hippocampal volume loss in Alzheimer’s patients compared to controls.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=58#h5p-7
2. ANOVA Analysis: Conduct an ANOVA to evaluate the effect of Alzheimer’s on volume loss. This analysis will help determine if the volume loss is significantly different between Alzheimer’s patients and controls.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=58#h5p-8
3. ANCOVA Challenge: As an advanced challenge, use ANCOVA to evaluate the effect of Alzheimer’s on volume loss while controlling for the linear association between volume loss and initial volume. This analysis accounts for the initial volume of the hippocampus, providing a more nuanced understanding of the disease’s impact.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=58#h5p-9
This case study provides a practical application of MRI image processing and statistical analysis in R, demonstrating the power of these techniques in understanding complex neurological conditions like Alzheimer’s disease.
Functional Magnetic Resonance Imaging (fMRI) is a non-invasive neuroimaging technique that has revolutionized our understanding of the brain. It is primarily used to observe and measure brain activity, providing an invaluable tool for neuroscience research. fMRI operates on the principle that cerebral blood flow and neuronal activation are coupled. When a brain area is more active, it consumes more oxygen, and to meet this increased demand, blood flow to the active area also increases. This phenomenon is known as the hemodynamic response.
The raw data from fMRI scans are typically in the form of 3D images or volumes. These are acquired over time, resulting in a 4D dataset (3D space + time).
Preprocessing of fMRI data is essential for enhancing data quality, standardizing data across sessions and subjects, removing artifacts due to subject movement and physiological processes, and optimizing the interpretation of the BOLD signal, thereby ensuring accurate and reliable subsequent analyses.
In this section, we will focus on how you can view and work with fMRI data in R.
While fMRI data can be saved in raw formats like DICOM or scanner-specific PAR/REC, the most common types for analysis are processed formats like the widely used NIfTI, which stores brain volume data and information, and BIDS, a standardized directory structure facilitating data sharing and compatibility with various analysis tools. Choosing the right type depends on analysis stage and compatibility needs, but NIfTI and BIDS are generally preferred for processed data due to their flexibility and widespread adoption. For this tutorial we use NIFTI file formats.
From here you can download a sample fMRI data saved in NIFTI.
1. Installing and Loading Necessary Packages:
install.packages(c("oro.nifti", "fmri", "neurobase", "fslr"))
library(oro.nifti)
library(fmri)
library(neurobase)
library(fslr)2. Loading fMRI Data:
# Load a NIfTI file
fmri_data <- readNIfTI("path/to/your/fmri.nii")
# Check the dimensions and structure
dim(fmri_data) # Check dimensions (x, y, z, time)
# As you can see fMRI has 4 dimensions which are 3D space + time.
str(fmri_data) # View data structure3. Visualizing fMRI Data:
# Display a single slice
ortho2(fmri_data, xyz = c(40, 40, 20)) # Visualize slice at coordinates (40, 40, 20)In fMRI, a voxel is a tiny 3D brain chunk like a pixel in an image. It measures blood flow changes linked to brain activity, giving us a detailed picture of what’s happening where and when.
# Extract time series from a specific voxel.
voxel_time_series <- fmri_data[25, 30, 15, ]
# Plot the time series
plot(voxel_time_series, type = "l", xlab = "Time", ylab = "BOLD Signal")In a neuroimaging study, a fMRI experiment was conducted to investigate brain responses to visual stimuli. The study employed a block design paradigm where participants were shown images of a baby at fixed intervals. Specifically, each participant was exposed to a picture of a baby for 15 seconds, followed by a 15-second interval where no image was displayed (black screen). This sequence was repeated for 10 cycles, resulting in a total experiment duration of 300 seconds, or 5 minutes.
The accompanying dataset voxels.csv contains time series data from 10 selected brain voxels of one participant, providing a focused insight into localized brain activity during the experiment. The data is structured to facilitate analysis of brain response patterns in relation to the visual stimulus. Additionally, the dataset includes a stimuli column that chronicles the timing of the visual stimuli presented to the participant. In this column, a value of 1 denotes the presence of the baby image on the screen, while a 0 indicates a phase where no image was shown (black screen). The fMRI data has been recorded every 1 second therefore there are 300 time-steps in the data.
Certainly! Here are some questions that you could ask students related to each part of the provided R code, aimed at testing their understanding of data analysis and visualization in R:
voxels_data. Explain what each part of the command does.voxels_data in R? Why is this step important before proceeding with data analysis?voxels_data? What kind of information does this function provide?apply function is used in this context.cor() function do? In this specific context, what are we trying to find out by using cor()?For more detailed information and advanced techniques in statistical analysis of fMRI using R, the following resources are recommended:
Have you ever wondered how to make sense of a dataset? Sometimes datasets are available but in formats that seem confusing? Well, sometimes, the organizational format in which you receive a dataset might not make very much sense or be of much value to tell you much about the data. To gain insights from your data, sometimes you need to make use of data organization tools – in R, we call this series of tools data wrangling.
Loading the tidyverse series of packages – a group of packages (tidyr, dplyr, and ggplot2) we can readily organize, tidy and visualise our data to check that our data are being organized into sensible formats.
In this markdown, we’ll be taking a look through some of the core tidyverse operations that are among the most commonly used, and use them to wrangle a categorical dataset to succinctly present information.
A few handy resources : here are some core “cheatsheets” for data cleaning and wrangling in RStudio using the tidyverse series of packages (cheatsheets developed by posit) :
Data Wrangling : https://www.rstudio.com/wp-content/uploads/2015/02/data-wrangling-cheatsheet.pdf
Tidyr : https://bioinformatics.ccr.cancer.gov/docs/rintro/resources/tidyr_cheatsheet.pdf
Dplyr : https://nyu-cdsc.github.io/learningr/assets/data-transformation.pdf
library(tidyverse) # core series of data wrangling packages.
library(dplyr) # core data wrangling grammar
library(ggplot2) # data visualisation tools
library(RColorBrewer) # colour palettes
library(here) # file directories
library(gridExtra) # arranging plots The dataset for this series of tidyverse exercise come from the UC Irvine Machine learning dataset repository. Summary information about the dataset and variables therein can be found at the following link : https://archive.ics.uci.edu/dataset/915/differentiated+thyroid+cancer+recurrence
Further reading about the dataset and use can be found in the source paper :
Borzooei, S., Briganti, G., Golparian, M. et al. Machine learning for risk stratification of thyroid cancer patients: a 15-year cohort study. Eur Arch Otorhinolaryngol (2023). https://doi.org/10.1007/s00405-023-08299-w
Load a copy of the thyroid cancer dataset and print the head (first 6 rows) of the dataset
thyroid.data <- read.csv("Thyroid_Diff.csv")
print (head(thyroid.data))
export.path <- here::here("/Tidyverse_DataWrangling")dataframes have a defined structure to them and there’s a certain terminology used to describe the different structures dataframes can take on.
If you view the thyroid cancer dataframe – is the dataframe tidy? Is the dataframe long or wide?
for more information on data wrangling and operations across the tidyverse, you can consult this chapter from the online book R for Data Science : 2e (information from which was used in constructing these activities) : https://r4ds.hadley.nz/data-transform
view(thyroid.data)
# the dataframe is tidy and in a wide format - wrangling will be needed to present informative counts from within the dataframe Now when you looked at the thyroid.data dataframe – every column describes something about each patient and their associated thyroid cancer. But how do you make sense of trends or readily visualise information in the dataframe? To do this, you’ll need to a little of what we call data WRANGLING. The process of organising and modifying the structure of your dataframe to present readily relevant information through statistics or succinct, targeted visualisations.
If you print the column names you see there are lots of different categorial features that describe the patient and their cancer. How can you visualise categorical data in a way that presents counts or proportions based on these categorical variables? This data wrangling activity will help you do just that. Into the tidyverse! 🙂
colnames ( thyroid.data)The first thing to understand how to use is the pipe operator %>%. This little data wrangling champion allows you to cleanly write and “daisy chain” a series of data handling and wrangling operations to seamlessly carry out a series of data manipulations to produce the desired structure of dataframe with the requisite columns and rows needed to produce the visualisations that best present the information contained therein.
First, the pipe operator can be applied to vectors as well as dataframes. In the same way we can “pipe” outputs of vectors into operations, we can “pipe” columns or entire dataframes into data wrangling functions to produce a required data structure.
to start for the next series of questions you will need to be comfortable using the pipe operator to wrangle your data from the thyroid cancer dataframe you loaded into RStudio. the first activity involves wrangling the dataframe to count how many thyroid cancer cases there are for each of the 4 different pathologies in the dataset.
## ------------------------------------------------------- ##
#### Using the pipe operator to pass inputs to functions ####
## ------------------------------------------------------- ##
# give this code and make it visible to the students
# try computing the mean of a vector of numbers :
no.piped.mean <- mean(c(1,2,3,4,5,6,7,8,9, 10, 11, 12))
piped.mean <- c(1,2,3,4,5,6,7,8,9,10,11, 12) %>% mean()
print (paste("mean without pipe operator: ", no.piped.mean,
"mean with pipe operator: ", piped.mean))
## --------- wrangling the thyroid cancer dataframe ---------- ##
## --------------------------------------------- ##
##### 1. grouping thyroid cancer by pathology #####
## --------------------------------------------- ##
# this is the first step to organizing the dataframe for downstream analysis
# the pipe operator is taking the thyroid.data dataframe and applying the group_by function to it grouping rows together based on the values in the pathology column of the dataframe.
thyroid.by.pathology <- thyroid.data %>% group_by(Pathology)
print (head(thyroid.by.pathology))
## ----------------------------------------------- ##
##### 2. summarise the counts of each pathology #####
## ----------------------------------------------- ##
# now you can prepare a frequency table - of all the cases in this thyroid cancer dataset, how many cases are of EACH pathology?
# this step "pipes" the thyroid.by.pathology dataframe produced in the previous step, to the summarise function, counting the frequency of each pathology across the entire dataset
pathology.frequencies<- thyroid.by.pathology %>% summarise (Frequency = n())
print (head(pathology.frequencies))There are 4 unique cancer pathologies in this dataset – Follicular, Hurthel Cell, Micropapillary, and Papillary. How many cases of each type are in this dataset? Present your results as a frequency histogram. Annotate the bars of the frequency histogram to show the number of cases in each pathology.
Hint : use the tidyverse series of packages to wrangle your data
# Use dplyr to group by Pathology and count the number of occurrences
# this time all the steps outlined in the preliminary code chunk was piped together and brought together to produce a pathology frequency dataframe.
pathology_freq <- thyroid.data %>%
group_by(Pathology) %>%
summarise(Frequency = n())
# Print the frequency distribution
print(pathology_freq)
# Use ggplot2 to create a bar plot of the frequency histogram.
thyroid.cancer.freqplot <- ggplot(pathology_freq, aes(x = Pathology, y = Frequency)) +
geom_bar(stat = "identity", fill = "lightsteelblue3") +
geom_text(aes(label = Frequency), vjust = 1.5, hjust = 0.5) +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
theme_classic()+
labs(x = "Pathology", y = "Frequency", title = "Frequency Distribution : Pathology Categories")
# plot inspection
print (thyroid.cancer.freqplot)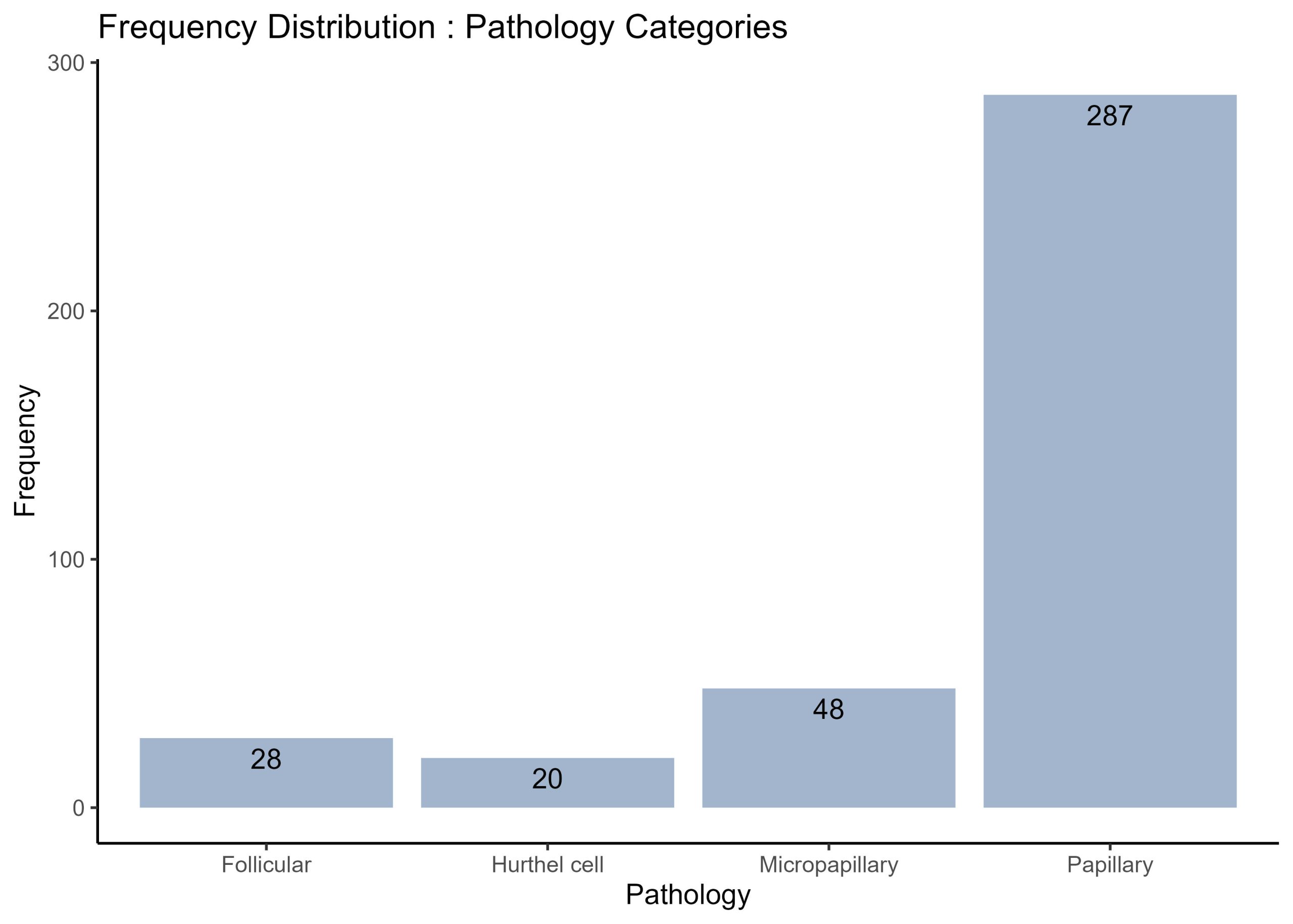
This first plot of a frequency histogram is nice because it readily visualises how many cases of each type of cancer pathology are in the dataset, but it’s important to also note that there are additional patient data that are also valuable, but not present in the previous visualisation. Supposed you would like to also know how many males and females there are for each pathology. How woud you obtain the proportions of males and females for each pathology and modify the frequency histogram above to visualise the counts of males and females for each pathology group?
Modify the frequency histogram above to show the proportions of males and females in each pathology, and annotate each bar with the counts of males and females in each pathology group.
# Group by Pathology and Gender, and count the number of occurrences
gender_pathology_freq <- thyroid.data %>%
group_by(Pathology, Gender) %>%
summarise(Frequency = n())
# Plot the stacked bar chart
thyroid.freqplot.by.Gender <- ggplot(gender_pathology_freq, aes(x = Pathology, y = Frequency, fill = Gender)) +
geom_bar(stat = "identity") +
geom_text(aes(label = Frequency), position = position_stack(vjust = 0.5)) +
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
theme_classic()+
labs(x = "Pathology", y = "Frequency", fill = "Gender", title = "Frequency Distribution of Each Pathology Category by Gender") # set the fill of each bar to show the proportions of genders
# plot inspection step
print (thyroid.freqplot.by.Gender)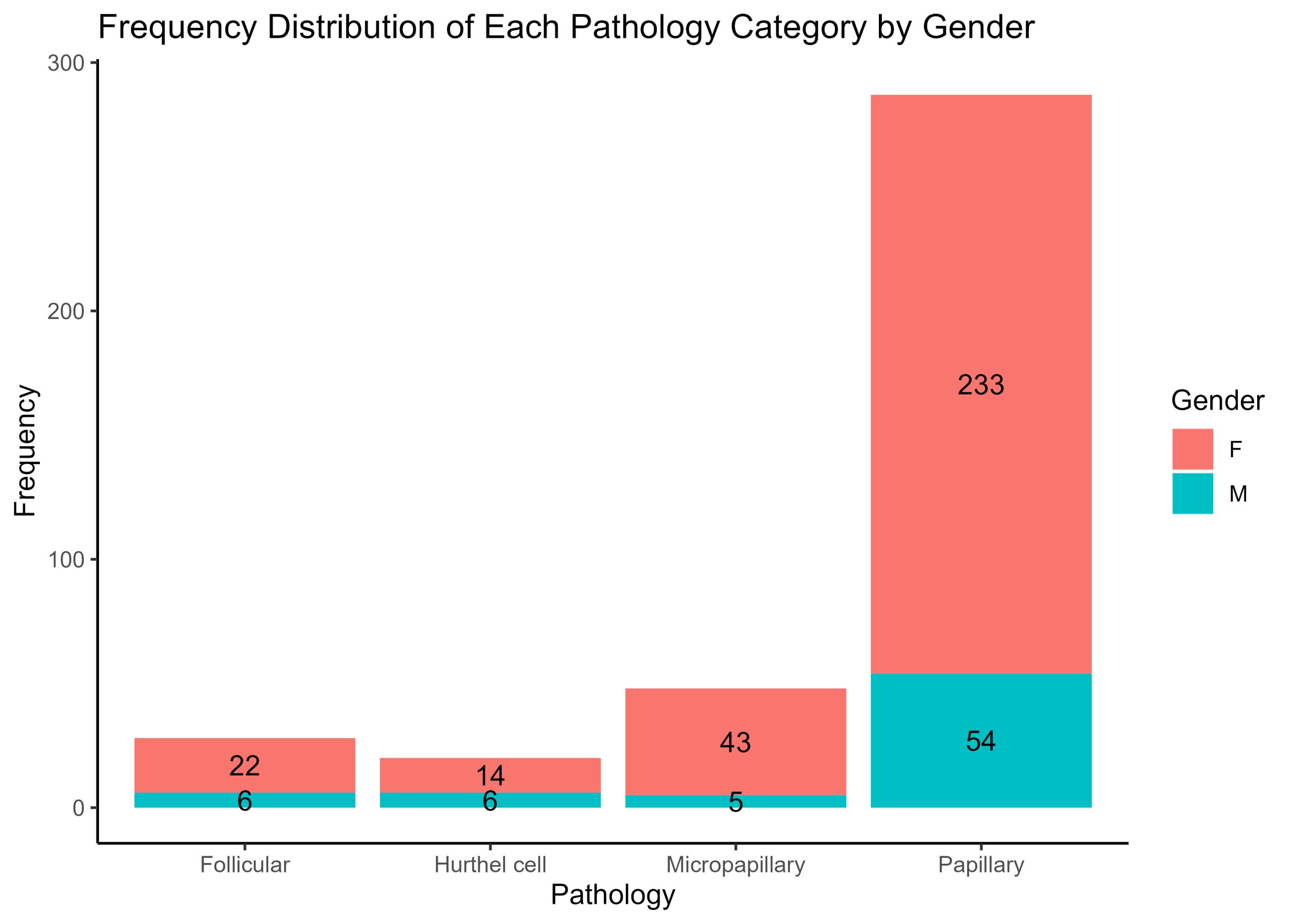
Frequency histograms are a useful way to visualise counts and prorportions, but suppose you would like to see the proportion of one set of diagnostic conditions across a series of cancer pathologies. For each patient, physical examination of the thyroid gland has been categorised into one of 5 options : diffuse goiter, multinodular goiter, normal, single nodular goiter – left, and single nodular goiter-right.
There are four distinct pathologies (as you know from the previous exercise). Suppose you are making sense of these data and want to see the prorportions of each physical diagnosis category across each of the pathologies. Yes, you could use a frequency histogram as developed previously, but alternatively you could use a pie chart to make proportions of physical diagnostic categories readily visible.
Prepare a series of 4 pie charts – one pie chart for each pathology and in each pie chart show the proportion of cases for each physical examination category.
#create a custom colour palette :
colour.palette <- c("maroon3", "mediumslateblue", "olivedrab3", "cadetblue2", "darkgoldenrod2" )
# Group by Pathology and Physical.Examination, and count the number of occurrences
pathology_exam_freq <- thyroid.data %>%
group_by(Pathology, Physical.Examination) %>%
summarise(Frequency = n())
# Calculate the total number of each Pathology
total_pathology <- pathology_exam_freq %>%
group_by(Pathology) %>%
summarise(Total = sum(Frequency))
# Join the two dataframes together
pathology_exam_freq <- left_join(pathology_exam_freq, total_pathology, by = "Pathology")
# Calculate the proportion
pathology_exam_freq <- pathology_exam_freq %>%
mutate(Proportion = Frequency / Total)
# Create a pie chart for each Pathology - to see the distribution of physical examination values
Diagnosis.by.Pathology <- pathology_exam_freq %>%
ggplot(aes(x = "", y = Proportion, fill = Physical.Examination)) +
geom_bar(width = 1, stat = "identity") + # fill each bar for a physical examination category
geom_text(aes(label = Frequency), position = position_stack(vjust = 0.5)) +
coord_polar("y", start = 0) + # create a circular histogram- pie chart
facet_wrap(~Pathology) + # this generates a series of 4 plots for each pathology
theme_void() + # aesthetics
theme(legend.position = "right") +
scale_fill_manual(values = colour.palette)+ # fill each bar with a specific colour from palette
labs(fill = "Physical Examination", title = "Proportion of Cases in Each Physical Examination Category by Pathology")
print (Diagnosis.by.Pathology)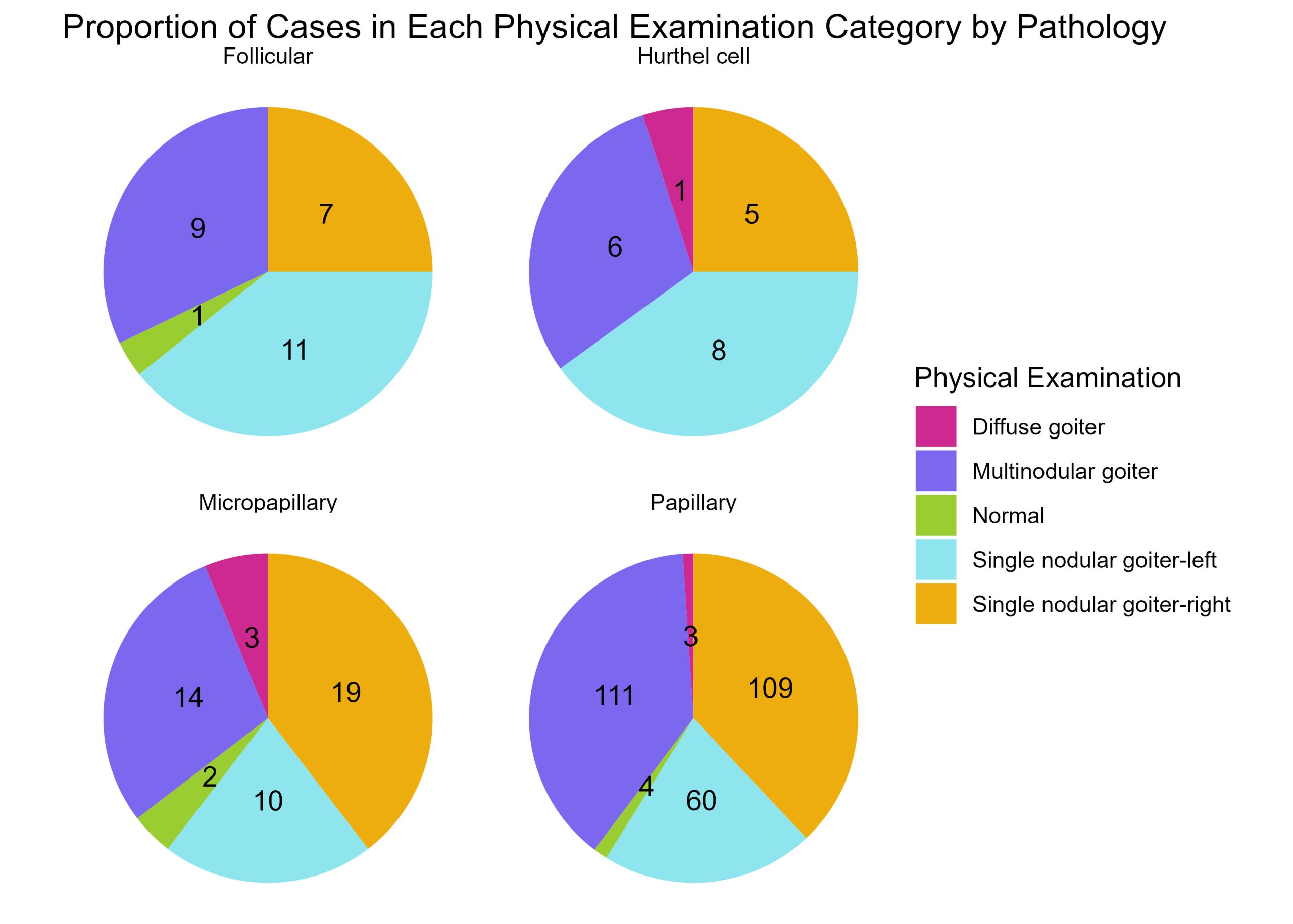
Now this is a nice way to show how many of each physical examination characteristics make up each cancer pathology. Now, just like you did for the frequency histogram above, separate these data to show the proportions of physical examination categories across each cancer pathology by Gender. This time, you need to create two series of 4 plots. One series for males and another for females. Each plot showing the proportion of physical examination categories for a given pathology.
#create a custom colour palette :
colour.palette <- c("maroon3", "mediumslateblue", "olivedrab3", "cadetblue2", "darkgoldenrod2" )
# Group by Gender, Pathology and Physical.Examination, and count the number of occurrences
gender_pathology_exam_freq <- thyroid.data %>%
group_by(Gender, Pathology, Physical.Examination) %>%
summarise(Frequency = n())
# Calculate the total number of each Gender and Pathology
total_gender_pathology <- gender_pathology_exam_freq %>%
group_by(Gender, Pathology) %>%
summarise(Total = sum(Frequency))
# Join the two dataframes together
gender_pathology_exam_freq <- left_join(gender_pathology_exam_freq, total_gender_pathology, by = c("Gender", "Pathology"))
# Calculate the proportion
gender_pathology_exam_freq <- gender_pathology_exam_freq %>%
mutate(Proportion = Frequency / Total)
# Create a pie chart for each Gender and Pathology
Diagnosis.by.Pathology.by.Gender<- gender_pathology_exam_freq %>%
ggplot(aes(x = "", y = Proportion, fill = Physical.Examination)) +
geom_bar(width = 1, stat = "identity") +
geom_text(aes(label = Frequency), position = position_stack(vjust = 0.5)) +
coord_polar("y", start = 0) +
facet_grid(Gender ~ Pathology) + # create a grid of plots - gender = column, pathologies = rows
theme_void() +
theme(legend.position = "top",
plot.title = element_text(hjust = 0.5)) +
scale_fill_manual(values = colour.palette)+
labs(fill = "Physical Examination", title = "Proportion of Cases for Each Physical Examination Category by Pathology and Gender")
print (Diagnosis.by.Pathology.by.Gender)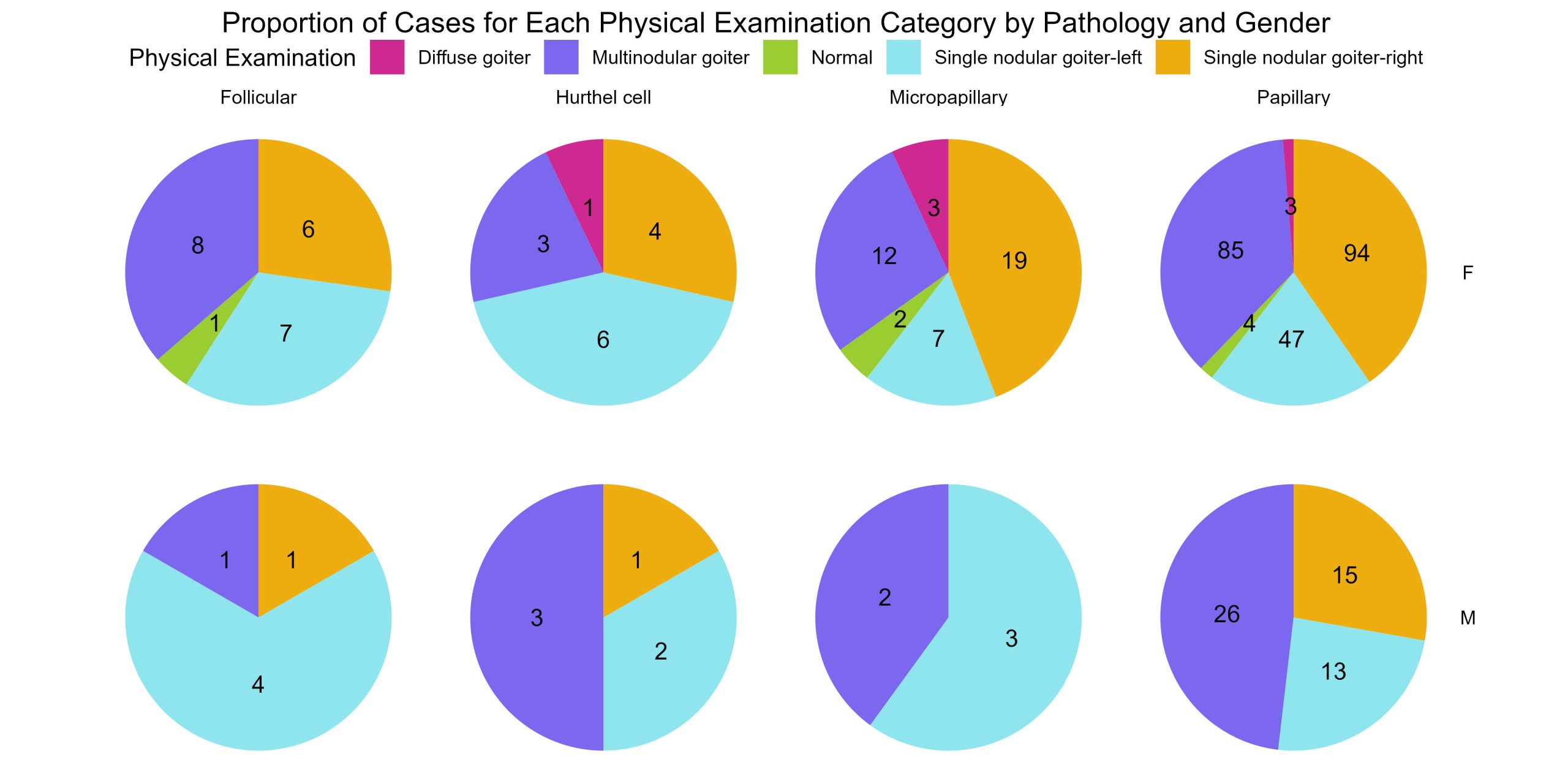
The data wrangling grammar given by core packages in the tidyverse provide a crucial way to group and organise your data to visualise trends, counts, and proportions more simply with compelling, succinct visualisations.
A prime example of this would be to create a series of boxplots. The series of 8 piecharts you just generated in the last exercise show the proportions of physical examination categories for each pathology for each Gender. But every patient in this dataset is not the same age. It would be important to know age demographics as well. Create a series of boxplots showing the age by gender for each physical examination category for each cancer pathology. This is a series of 16 boxplots.
This time we need to use boxplots instead of piecharts or frequency histograms because we want to show the range of ages by gender for each group of categories.
# Create boxplots of Age by Gender for each Physical.Examination for each Pathology
thyroid.age.boxplots <- thyroid.data %>%
ggplot(aes(x = Gender, y = Age, fill = Gender)) +
geom_boxplot() +
facet_grid(Pathology ~ Physical.Examination) + # produces a grid of plots - each row is a pathology, and each column is a physical examiantion category with boxplots of age by gender
theme(axis.text.x = element_text(angle = 45, hjust = 1)) +
labs(x = "Gender", y = "Age", fill = "Gender", title = "Boxplots of Age by Gender for Each Physical Examination Category by Pathology")
print (thyroid.age.boxplots)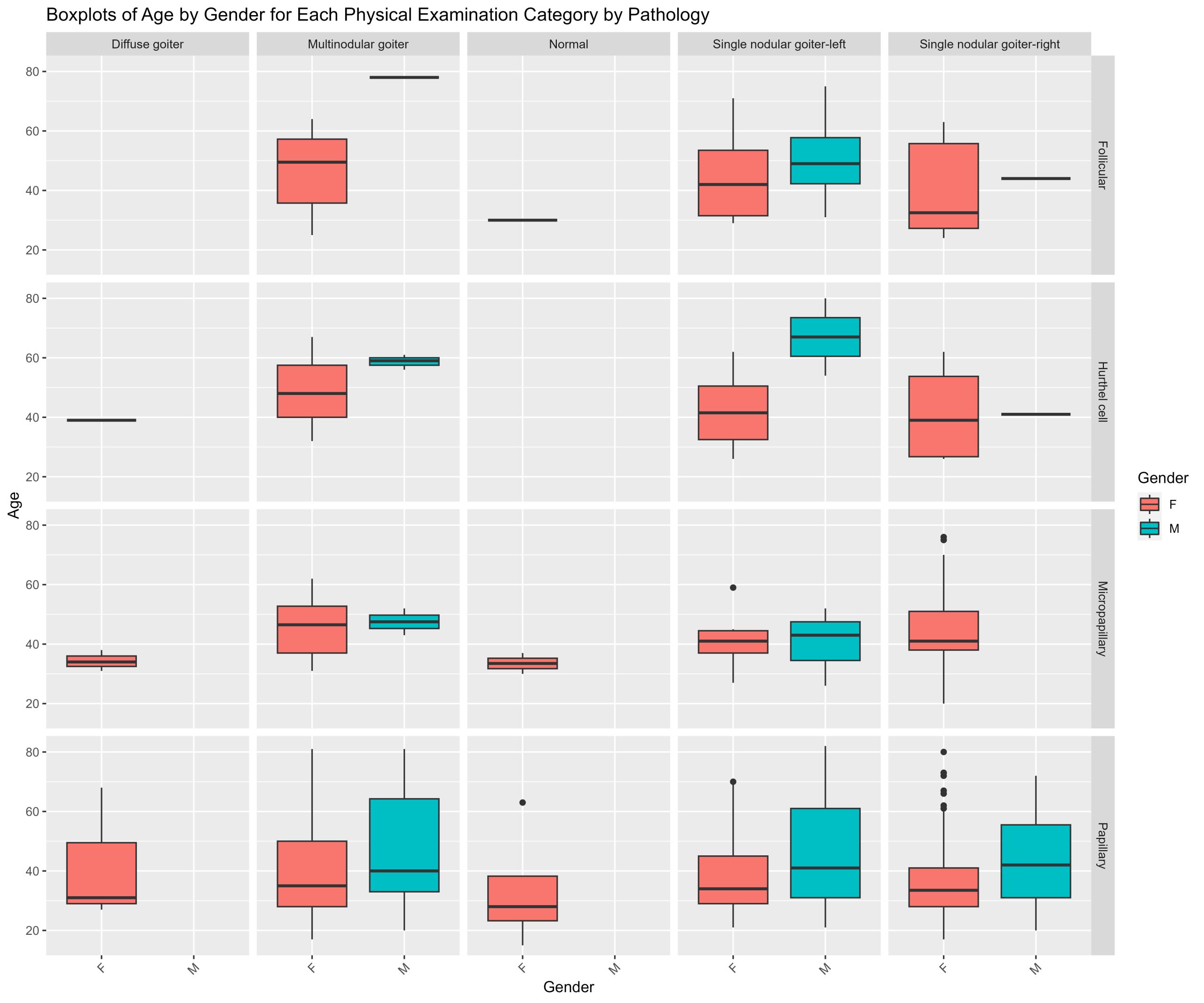
Although you could prepare a series of counts, prorportions or boxplots for various combinations of features nested together. One critical step is to understand what your features are, what the categorical information contained in each feature means, and what proportions of your data are found across each feature. You can accomplish this by creating pie charts for every feature except for patient age. Though you would probably perform this at the beginning before you undertake your own analysis of a categorical dataset, this process is a little more complicated so we’re saving it for last. The process involves both a series of wrangling operations and visualisation tools.
It’s from exploratory analyses like the plots generated in this grid of pie charts that chaining feature information from pathology, gender, physical examination, and age seemed interesting!
# Exclude the "Age" column
data_without_age <- thyroid.data[ , !(names(thyroid.data) %in% "Age")]
# Initialize an empty list to store the plots
plot_list <- list()
## --------------------------------------------- ##
###### loop through columns - get proportions #####
###### & Generate the pie charts each column #####
## --------------------------------------------- ##
for (column_name in names(data_without_age)) {
# Calculate proportions
proportions <- data_without_age %>%
group_by(.data[[column_name]]) %>% # group by common features.
summarise(n = n()) %>% # produce summary statistics
mutate(prop = n / sum(n)) # mutate produces a column with proportions in one step
# Create pie chart
pie_chart <- ggplot(proportions, aes(x = "", y = prop, fill = .data[[column_name]])) +
geom_bar(width = 1, stat = "identity") +
coord_polar("y", start = 0) +
labs(title = paste("Pie Chart for", column_name), x = NULL, y = NULL, fill = column_name) +
theme(plot.margin = margin (1,1,1,1, "cm"))+ # common margin to each plot
theme_void() #aesthetics - blank backgrounds
# Add the pie chart to the list
plot_list[[column_name]] <- pie_chart
}
# Arrange the plots into a grid
plotgrid <- grid.arrange(grobs = plot_list, ncol = 4, returnGrob= TRUE)
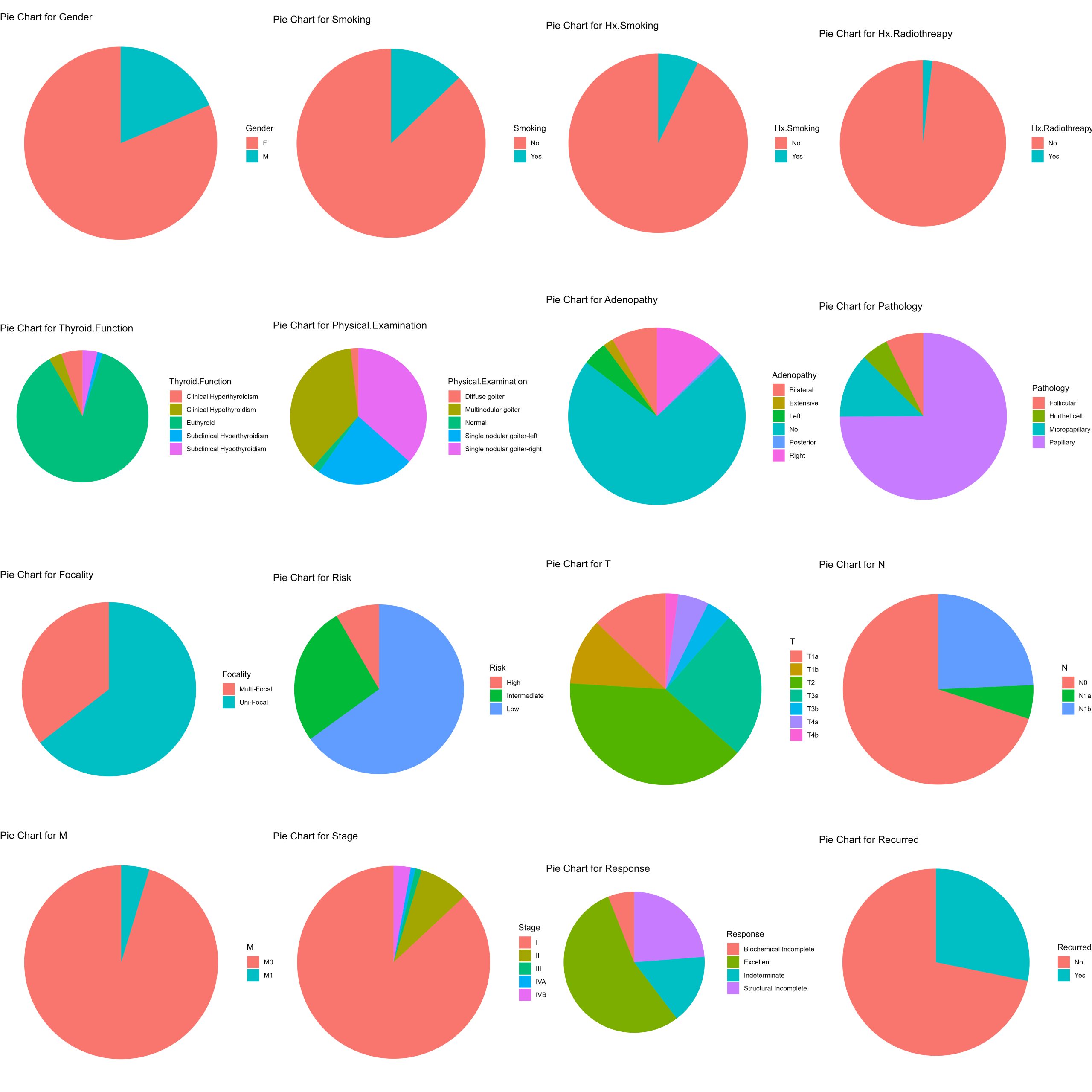
To download, right-click and press “Save File As” or “Download Linked File”
You are a pathologist and have taken the measurements of 569 nuclei of cells from needle aspirates of breast tissue masses. Samples come from either benign (B) or malignant (M) masses. You would like to perform an analysis of the cell shapes and sizes between the malignant and benign cells to better understand the differences between them. You would also like to explore using machine learning to see how well simple clustering algorithms can identify benign cells from malignant ones based SOLELY on their size and shape features. Using the Wisconsin Breast Cancer Dataset
———– Text from Kaggle ——————– The breast cancer data includes 569 examples of cancer biopsies, each with 32 features. One feature is an identification number, another is the cancer diagnosis and 30 are numeric-valued laboratory measurements. The diagnosis is coded as “M” to indicate malignant or “B” to indicate benign.
The other 30 numeric measurements comprise the mean, standard error and worst (i.e. largest) value for 10 different characteristics of the digitized cell nuclei, which are as follows:-
Radius Texture Perimeter Area Smoothness Compactness Concavity Concave Points Symmetry Fractal dimension ——————– Dataset Link Below ————— Breast Cancer Dataset available from : https://archive.ics.uci.edu/dataset/17/breast+cancer+wisconsin+diagnostic
for further reading on the Wisconsin Breast Cancer Dataset,specifically how each of the features in this dataset were calculated, please consult:
Street, W.N., Wolberg, W.H., & Mangasarian, O.L. (1993). Nuclear feature extraction for breast tumor diagnosis. Electronic imaging.
Install and load the following packages into R
library(dabestr) # estimation statistics
library(ggplot2) # plotting and data visualisation
library(pheatmap) # generating a heatmap
library(tidyverse) # data handling
library(dplyr) # data handling
library(stats) # basic statistics
library(RColorBrewer) # colour palettes and plot aesthetic controls
library(Rtsne) # for performing T-distributed stochastic neighbour embedding (tsne)
Download the dataset from the UCI Machine Learning Repository. Load the csv file of measurements into RStudio
BreastCancer.Data <- read.csv("WisconsinBreastCancerData.csv")
BreastCancer.Data$X <- NULL
print (head(BreastCancer.Data))The entire BreastCancer.Data dataframe is structured such that each row is a cell and each column is a parameter. However, there are certain columns that are not features (quantitative descriptions of the cells). When a column is not a feature, it’s a label. Labels are ways to identify or tag specific cells once we understand how they are characterised by their features.
Before we can explore the dataset, we need to separate the labels from the features.There are two columns in the dataframe that label the cells – id -> the unique identification number assigned to a cell – diagnosis -> whether the cell is from a benign (B) or malignant (M) Sample)
The remaining columns are features that serve as quantitative descriptions of the cell sizes and shapes
## ---------------------------------------- ##
##### Setting features apart from labels #####
## ---------------------------------------- ##
# labels dataframe - contains the unique identifier and the diagnosis)
Diagnostic.Labels <- select(BreastCancer.Data, id, diagnosis)
# features dataframe - all the columns that are not identifiers are the measured features that characterise the tissue mass - specifically the characterizing the nuclei present.
feature.columns <- setdiff (colnames(BreastCancer.Data), colnames(Diagnostic.Labels))
Diagnostic.Features <- BreastCancer.Data[, feature.columns]
# create a z-scored version of the features
Diagnostic.Features.Scored <- scale(Diagnostic.Features)
# print (head(Diagnostic.Features.Scored))
view(Diagnostic.Features.Scored)
print (colnames(Diagnostic.Features.Scored)Tsne or T-distributed Stochastic Neighbour Embedding is a technique used to visualise high-dimensional data in 2 dimensions. Termed a dimensionality-reduction method, it allows us to explore the data one feature at a time. In this breast cancer dataset, there are 32 measures that describe the size, shape, and texture of masses in breast tissue that then go on to be either benign (B) or malignant (M).
For further reading on T-sne, please consult : Van der Maaten, L., Hinton, G. Visualizing Data using T-sne. Journal of Machine Learning Research 9 (2008) 2579-2605
Let’s use tsne to visualise how these features are distributed across the dataset.
N.B. I ran this for loop of seed by perplexity combinations because it is important to test these parameters when wrorking with T-sne. The perplexity controls how much of the local vs. global data structure elements contribute to the final visualisation as the data pass through dimensionality reduction. Variations in perplexity for the same random seed can have very dramatic effects on the final visualisation despite the data not changing.
# Define your perplexities and seeds
perplexities <- c(10, 15, 20, 25, 30)
seeds <- c(123, 456, 789, 246, 135 )
# Create an empty dataframe to store the t-SNE components
tsne_data <- data.frame()
# Iterate over each combination of perplexity and seed
for (i in 1:length(perplexities)) {
for (j in 1:length(seeds)) {
# Perform t-SNE with the current perplexity and seed
tsne_model <- Rtsne(Diagnostic.Features.Scored, perplexity = perplexities[i], seed = seeds[j])
# Create a dataframe for the t-SNE components
tsne_temp <- data.frame(
tSNE1 = tsne_model$Y[, 1],
tSNE2 = tsne_model$Y[, 2],
Perplexity = perplexities[i],
Seed = seeds[j],
Diagnosis = Diagnostic.Labels$diagnosis
)
# Append the data to the main dataframe
tsne_data <- rbind(tsne_data, tsne_temp)
}
}
# Create the plot
tsne.stampcollection <- ggplot(tsne_data, aes(x = tSNE1, y = tSNE2, colour = Diagnosis)) +
geom_point() +
facet_wrap(Perplexity ~ Seed, scales = "free") +
theme_bw()
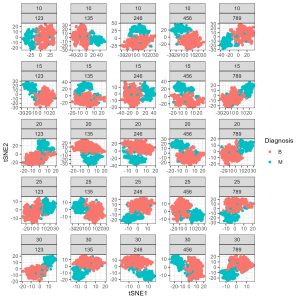
Use the following parameters in your initial tsne: – seed <- 789 – perplexity <- 30
Construct a T-sne map of the Diagnostic.Features dataframe. Plot the resulting T-sne map using ggplot making sure to colour the points by their diagnosis label. What do you notice about the T-sne map when it’s annotated by diagnosis?
As an exercise – vary the perplexity for the seed of 789, try ranging from 10 to 50 by skips of 10. What do you notice about the T-sne map as you vary the perplexity? What do you think the perplexity parameter controls in the T-sne algorithm?
# Set your perplexity and seed
set.seed (789)
perplexity <- 30
seed <- 789
# Perform t-SNE with the specified perplexity and seed
tsne_model <- Rtsne(Diagnostic.Features.Scored, perplexity = perplexity, seed = seed)
# Create a dataframe for the t-SNE components
tsne_data <- data.frame(
tSNE1 = tsne_model$Y[, 1],
tSNE2 = tsne_model$Y[, 2],
Diagnosis = Diagnostic.Labels$diagnosis # Add the diagnosis column
)
# Load ggplot2
library(ggplot2)
# Create the plot
tsne.by.diagnosis <- ggplot(tsne_data, aes(x = tSNE1, y = tSNE2, colour = Diagnosis)) +
geom_point() +
labs( x = "T-SNE Component 1", y= "T-SNE Component 2")
theme_bw()
# Print the plot
print(tsne.by.diagnosis)
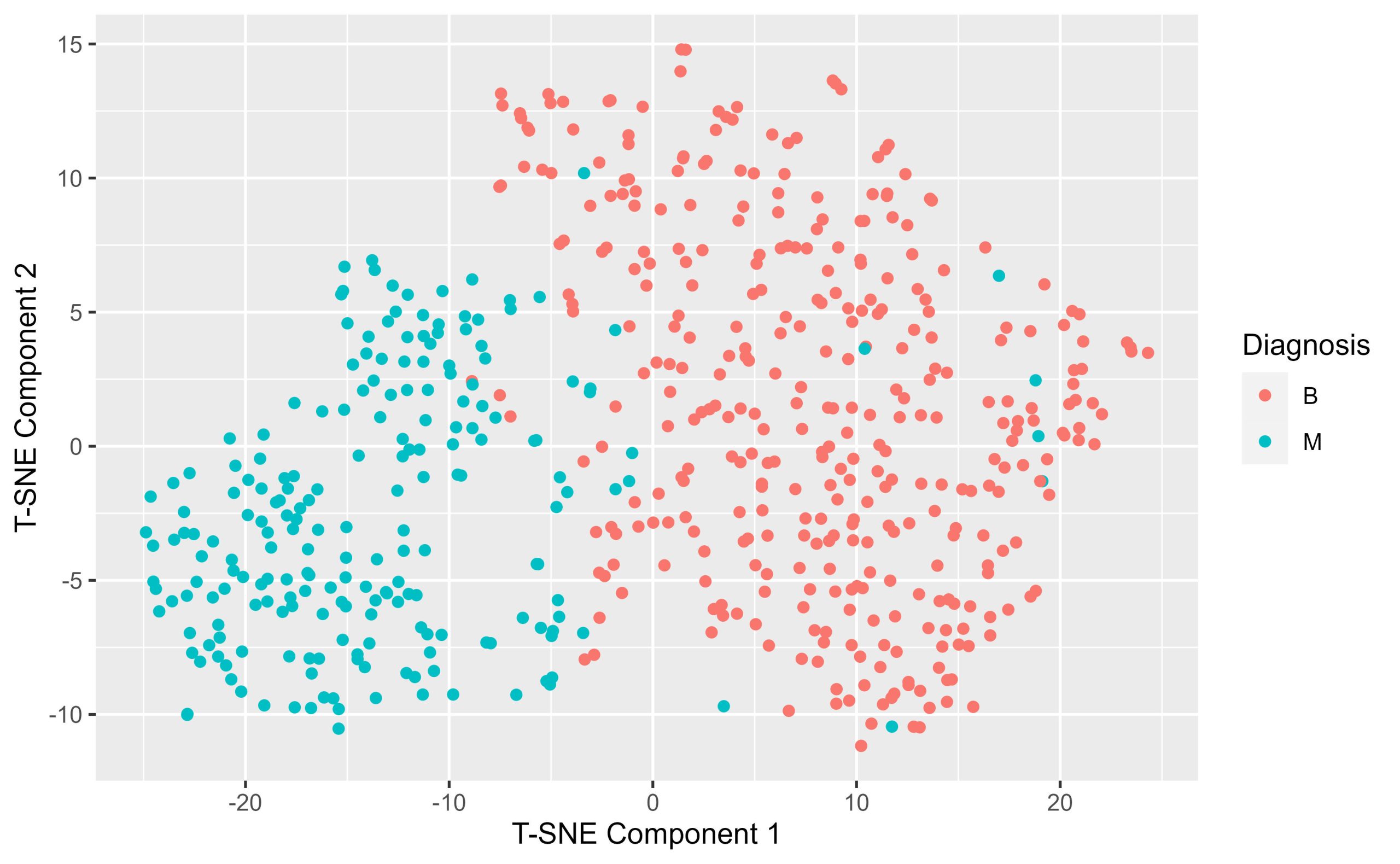
What if there were a way to readily identify cells as being benign or malignant depending on their size and shape parameters? Clustering algorithms are an example of a data-driven approach to grouping your data together based on the patterns present in the features. Of course, the data contain labels, but what if you clustered on the feature values, the measurements, without providing any information about the cell diagnosis? Select 2 clusters – we want to see whether hierarchical clustering separates benign vs malignant cells based on their shapes and size information only.
The steps to performing hierarchical clustering are as follows : 1.construct a dissimilarity matrix of the z-scored features. Use Euclidean Distances 2.call the hclust function on the dissimilarity matrix and specify the method is ward.D2 3.annotate the dendrogram to show where the data are partitioned into clusters 4. visualise the results of your clustering by constructing a heatmap of the dissimilarity matrix organised with an annotated surrounded dendrogram.
## ----------------------------------------------------- ##
#### dissimilarity matrix and hierarchical clustering #####
## ----------------------------------------------------- ##
# set the number of clusters
num.clusters <- 2
# construct a pairwise dissimilarity matrix using euclidean distances
dissimilarity.matrix <- dist(Diagnostic.Features.Scored, method = "euclidean")
# hierarchical clustering with Wthe ward.D2 algorithm
h.clusters <- hclust(dissimilarity.matrix, method = "ward.D2")
# cut the dendrogram into k clusters
clusters <- cutree(h.clusters, k = num.clusters)
## --------------------------------------------------- ##
##### Preparing Dataframe for Downstream Statistics #####
## --------------------------------------------------- ##
# create a dataframe of identifiers and z-scored features
All.Samples.zscored <- cbind(Diagnostic.Labels,
Diagnostic.Features.Scored)
# append the clusters to the dataframe
All.Samples.zscored$hclust_clusters <- clusters
# view(Malignant.Samples.zscored) # inspection step
##--------------------------------------- ##
##### Annotating the HClust Dendrogram #####
##--------------------------------------- ##
# create an annotation dataframe
annotation.df <- data.frame(Cluster = as.factor(clusters))
rownames(annotation.df) <- rownames(Diagnostic.Features)
# apply a colour palette to the annotations on the dendrogram
# colour palette for the k clusters
k.cluster.colourpalette <- c("olivedrab2", "maroon3")
# colour mapping
colourmapping <- setNames(k.cluster.colourpalette, levels (annotation.df$Cluster))
# make a list of annotation colours
annotation.colours = list(Cluster = k.cluster.colourpalette)
# match the names of the annotation colours to the cluster levels ( 1-8)
names(annotation.colours$Cluster) <- levels(annotation.df$Cluster)
## ----------------------------------------- ##
##### visualising clustering in a heatmap #####
## ----------------------------------------- ##
# set a colour palette
# diverging - spectral
div.spectral.red.blue <- c("#4a100e", "#731331", "#a52747", "#c65154", "#e47961", "#f0a882", "#fad4ac", "#ffffff",
"#bce2cf", "#89c0c4", "#5793b9", "#397aa8", "#1c5796", "#163771", "#10194d" )
div.spectral.blue.red <- rev(div.spectral.red.blue)
# interpolate the colours for continuous scales
continuous.spectral.redblue <- colorRampPalette(div.spectral.red.blue) (256)
continuous.spectral.bluered <- colorRampPalette(div.spectral.blue.red)(256)
# set the breaks in the colour scale
palette.breaks <- seq(from= 0, to = 8, length.out = length (continuous.spectral.redblue) + 1)
# generate the heatmap
hclust.dissimilarity.heatmap <- pheatmap(dissimilarity.matrix,
cluster_rows = h.clusters,
cluster_cols = h.clusters,
annotation_row = annotation.df,
# annotation_col = annotation.df,
annotation_colors = annotation.colours,
color = continuous.spectral.redblue,
breaks = palette.breaks)
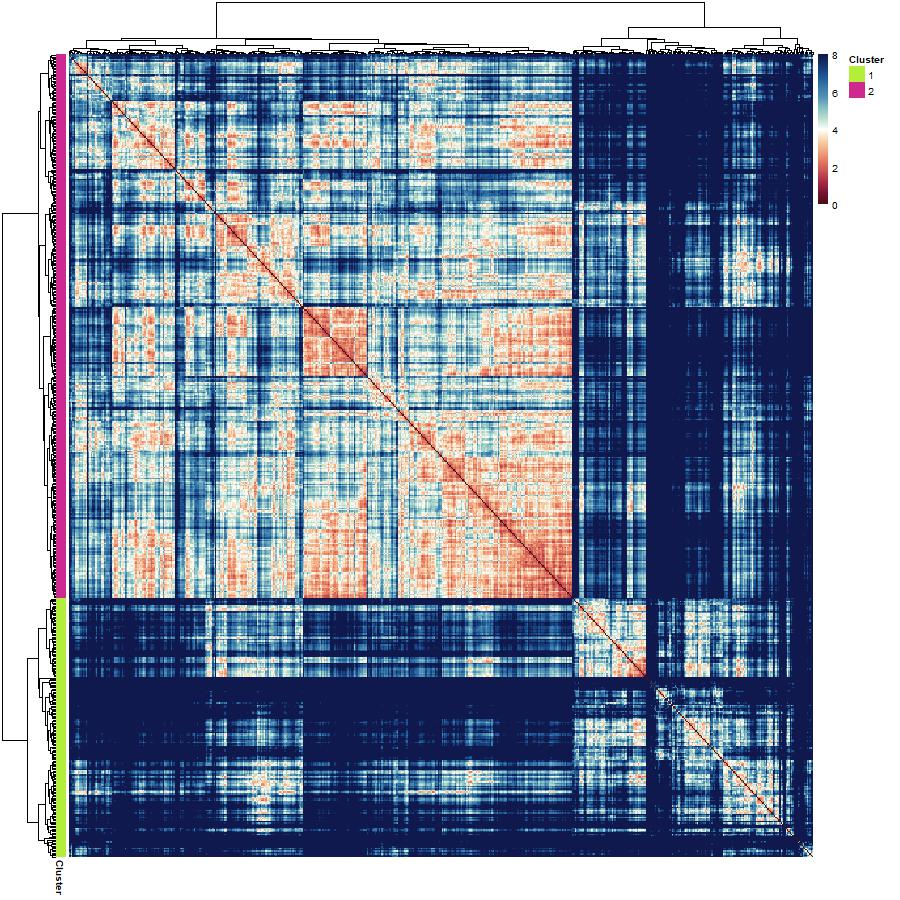
You have just produced a hierarchical clustering result. You can visualise the clustering with a heatmap with an annotated surrounding dendrogram, but that doesn’t tell you about which cells were relegated into which cluster. You can also produce a stacked proportion barplot that shows you the proportion of benign and malignant cells in a cluster.
library(dplyr)
# Calculate proportions of benign and malignant cells by cluster
# you can use the pipe operator to nest a series of operations to be performed to the dataframe of origin
All.Samples.zscored.grouped <- All.Samples.zscored %>%
group_by(hclust_clusters, diagnosis) %>%
summarise(n = n()) %>%
mutate(freq = n / sum(n))
view (All.Samples.zscored.grouped)
# Prepare a stacked proportion barplot
Hclust.barplot <- ggplot(All.Samples.zscored.grouped, aes(fill=diagnosis, y=freq, x=hclust_clusters)) +
geom_bar(position="fill", stat="identity") +
theme_minimal() +
labs(x="Cluster", y="Proportion", fill="Diagnosis") +
scale_x_continuous(breaks = c(1,2))+
theme(axis.text.x = element_text(angle = 90, hjust = 1))
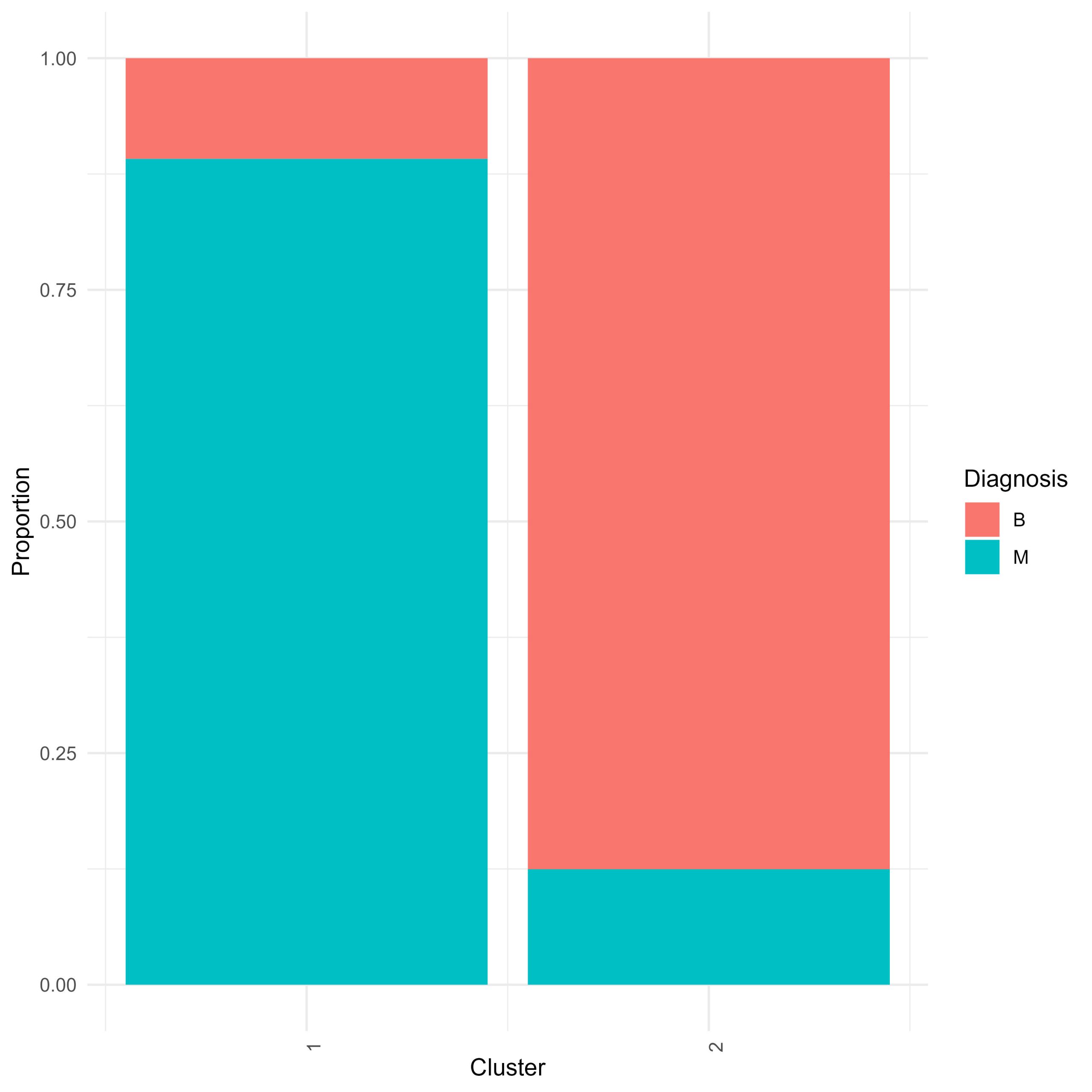
– Student Question – Across the features in your dataframe there are patterns in those data that can distinguish benign from malignant cells. You want to assess how different the benign and malignant cells are.
Estimation statistics are an alternative framework for statistical analysis that do not depend on p-values or significance testing. Instead they show the data, the distributions and their unpaired median differences along with bootstrapped 95% confidence intervals. This framework allows you to see your data and make assessments of difference/no difference without significance testing. It is a powerful framework that makes statistical inference visually accessible and frankly, beautiful!
The previous estimation statistics were applied to the cells categorised by diagnostic label – to assess for differences in features between benign and malignant cells. This time, repeat the same estimation statistics framework and look at the same features, but apply estimation statistics to the clusters instead. How do the estimation plots by cluster compare to the estimation plots by diagnostic label?
For further reading on estimation statistics please consult : Ho et al., 2019. published in Nature Methods
Ho, J., Tumkaya, T., Aryal, S. et al. Moving beyond P values: data analysis with estimation graphics. Nat Methods 16, 565–566 (2019). https://doi.org/10.1038/s41592-019-0470-3
# names of clusters
# cluster_names <- c("B", "M") # performing estimation stats by diagnostic label
cluster_names <- c("1", "2") # if performing estimation stats by hclust defined clusters
feature.of.interest <- "fractal_dimension_mean"
data1 <- All.Samples.zscored %>%
# select(variable = "diagnosis", value = feature.of.interest) # estimation on diagnostic labels
select (variable = "hclust_clusters", value = feature.of.interest) # estimation on the hclust clusters
estimation.stats.data <- data1
# Specify your reference group
# reference_group <- "B" # estimation by diagnostic label ( B = Benign, M = Malignant)
reference_group <- "1" # estimation by hclust cluster
# set the control group
control = reference_group
# set the comparison groups
comparisons = setdiff(cluster_names, control)
# Set the column names
colnames(estimation.stats.data)[2] = "Z-Scored Mean Nuclear Fractal Dimension"
# Prepare data for estimations statistics processing
two.group.unpaired =
estimation.stats.data %>%
dabest(variable,`Z-Scored Mean Nuclear Fractal Dimension`,
idx = c(control, comparisons),
paired = FALSE) %>%
median_diff(reps = 10000)
# Set the color parameters
# colour palette corresponds to diagnostic labels
# colours = c("coral2", "turquoise3" )
# colour palette corresponds to hierarchically defined clusters
colours = c("maroon3", "olivedrab2")
colour.swarm.plot = c(colours[which(cluster_names == control)],
setdiff(colours,
colours[which(cluster_names == control)]))
swarm.plot <- plot(two.group.unpaired,
palette = colour.swarm.plot,
# tick.fontsize = 20,
# axes.title.fontsize = 25,
rawplot.type = "swarmplot",
rawplot.ylabel = "Z-Scored Mean Nuclear Fractal Dimension")+
ggtitle(paste("Reference: cluster ", control))+
theme(title = element_text(face = "bold"),
plot.title = element_text(hjust = 0.5, vjust = 10, size = 20,
margin = margin(t = 80, b = -35)))
print(swarm.plot)
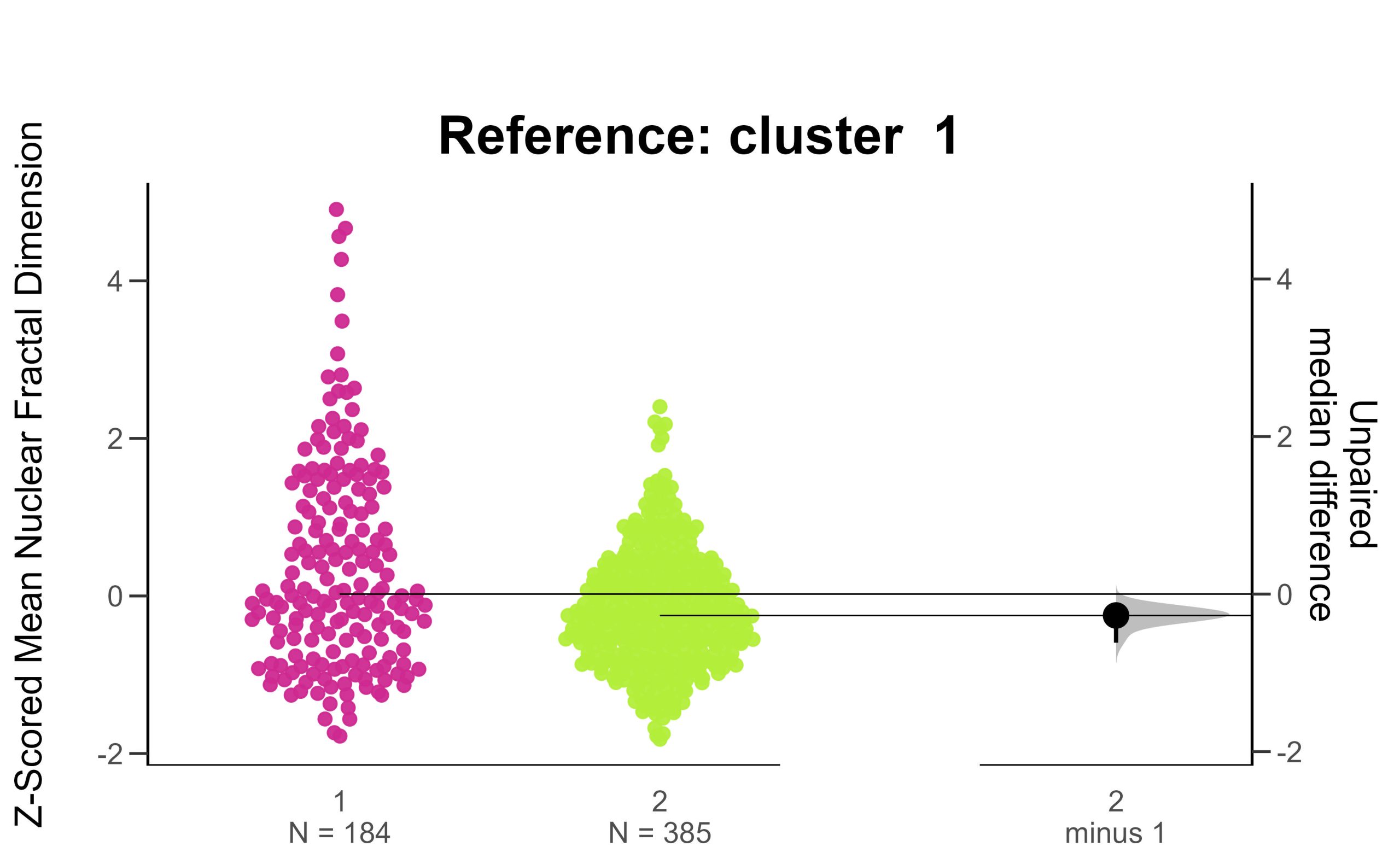
Another way to interrogate how well clustering assigned clusters based on diagnosis label is to annotate the T-SNE plot you constructed earlier but this time instead of colouring the points by their known diagnosis label, you can annotate by their hierarchically-defined clusters.
Use the same perplexity and random seed as before.This way you can compare how the hclust clusters map on to the high dimensional space against how the diagnostic labels map onto the same space. When comparing tsne maps annotated by diagnosis, and by hclust cluster, what do you notice? What similarities and differences stand out to you?
## --------------------------------------------------- ##
##### Repeat T-SNE code but plot coloured by hclust #####
## --------------------------------------------------- ##
# Set your perplexity and seed
set.seed (789)
perplexity <- 30
seed <- 789
# Perform t-SNE with the specified perplexity and seed
tsne_model <- Rtsne(Diagnostic.Features.Scored, perplexity = perplexity, seed = seed)
# Create a dataframe for the t-SNE components
tsne_data <- data.frame(
tSNE1 = tsne_model$Y[, 1],
tSNE2 = tsne_model$Y[, 2],
hclust_cluster = as.factor(All.Samples.zscored$hclust_clusters) # Add the Hierarchical Cluster Assignment - convert to factor to make the scale discrete
)
view( tsne_data)
# Load ggplot2
library(ggplot2)
# specify the colour palette :
hclust.colours <- c("maroon3", "olivedrab2")
# Create the plot
tsne.by.hcluster <- ggplot(tsne_data, aes(x = tSNE1, y = tSNE2, colour = hclust_cluster)) +
geom_point() +
labs( x = "T-SNE Component 1", y= "T-SNE Component 2") +
theme_bw() +
scale_color_manual(values = hclust.colours)
# Print the plot
print(tsne.by.hcluster)
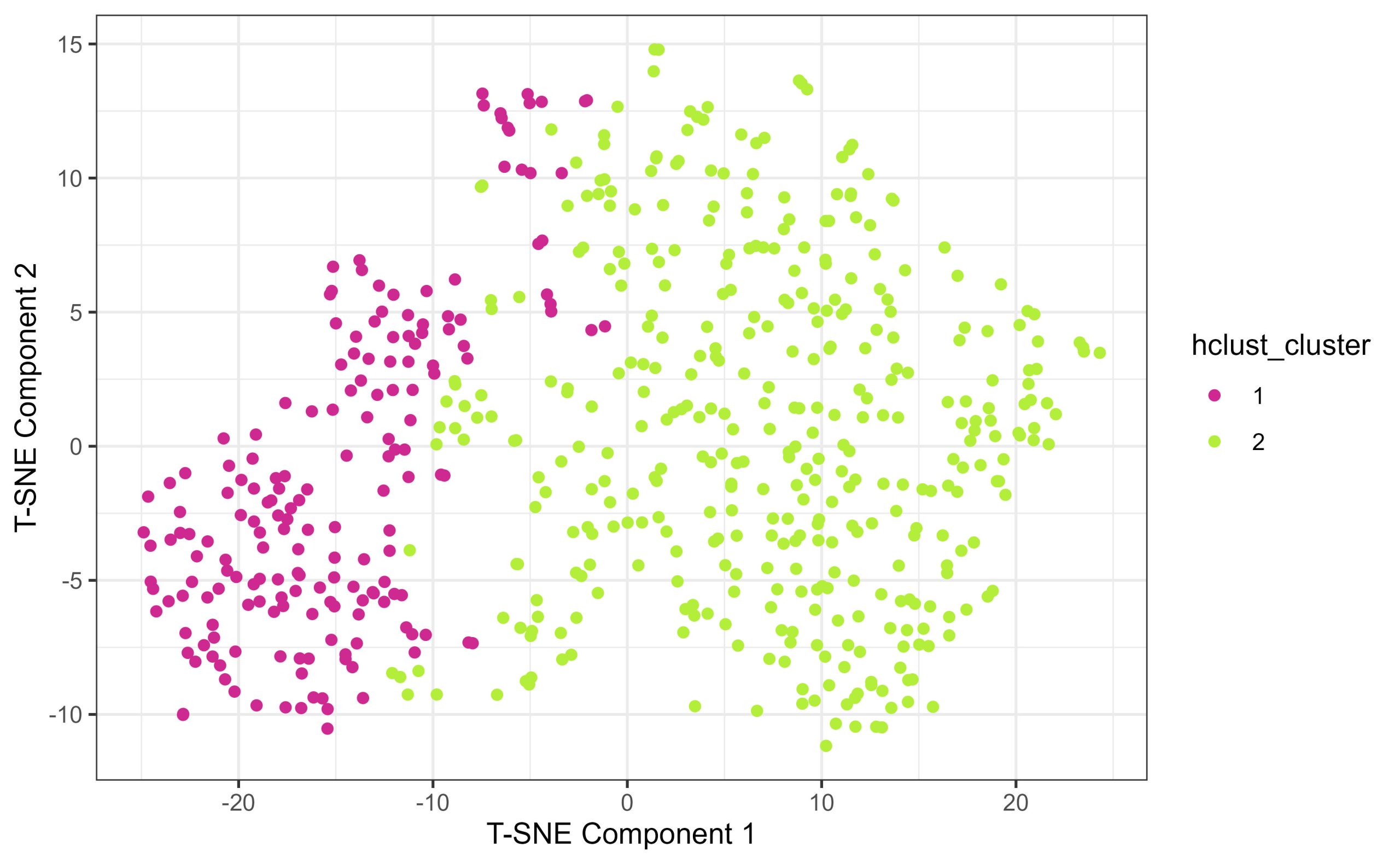
(something for students to think about – can be made into a discussion about clustering algorithms, and why it’s important to verify and validate clustering outputs)
You compared the true diagnostic labels for breast cancer cells – benign and malignant to hierarchical clusters constructed solely from size and shape features. Based on how well the clustering algorithm separated benign and malignant cells, think about how useful this approach would have been if you did not know a priori that the cells were classified into benign and malignant diagnostic categories. After you clustered them into two groups and compared their statistics, what would you need to do to confirm the cells of one cluster being malignant and the others are benign?
III
You are an evolutionary biologist studying sexual conflict and the effect of mating frequency on female lifetime fitness. Your study system is the notorious bedbug Climex lectularis. Bedbugs live in mixed-sex aggregations where females are subject to frequent reproductive harassment from males. Due to this reproductive harassment, females vary in their mating rates in natural groups where some females mate relatively infrequently, and others mate much more frequently. Higher mating rates for females increase the number of offspring produced per unit time, but may impose a longevity penalty as the physical process of reproduction is costly. This trade-off begs the question of whether variation in mating rate leads to a change overall lifetime fitness (number of viable offspring produced). To answer this question, you subjected females to a high-frequency (High) or low-frequency (Low) mating frequency treatment (Treatment). You counted the number of total viable offspring each female produced in her lifetime (Hatchlings), as well as her total lifetime in days (Longevity). You recorded your results in the dataframe held in “femalefitness.csv”.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=79#h5p-11
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=79#h5p-12
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=79#h5p-13
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=79#h5p-14
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=79#h5p-15
To download, right-click and press “Save File As” or “Download Linked File”
Cognitive Ecology Lab, Dr. Reuven Dukas, McMaster University Department of Psychology, Neuroscience, & Behaviour https://psych.mcmaster.ca/dukas/index.htm
Yan, J. L., & Dukas, R. (2022). The social consequences of sexual conflict in bed bugs: social networks and sexual attraction. Animal Behaviour, 192, 109-117.
Parker, G. A. (2006). Sexual conflict over mating and fertilization: an overview. Philosophical Transactions of the Royal Society B: Biological Sciences, 361(1466), 235-259.
Maklakov, A. A., Bilde, T., & Lubin, Y. (2005). Sexual conflict in the wild: elevated mating rate reduces female lifetime reproductive success. the american naturalist, 165(S5), S38-S45.
You are behavioural ecologist interested in territoriality and aggression in frogs. The brilliant-thighed poison frog Allobates femoralis relies on the presence of small pools of water for their reproduction. Males of this species engage in territorial competition to secure areas that contain these pools of water. It is unknown, however, at which point in the frogs’ development the territorial behaviour emerges. To test this, you traveled to a field site in eastern Ecuador and captured 50 A. femoralis of various developmental stages. You recorded each frog’s sex, measured each frog’s Snout-Vent Length (SVL; in mm), then subjected the frogs to a mirror test. In the mirror test, you recorded whether they expressed aggressive behaviours (e.g. lunges, bites, or grappling) towards their reflections on the mirror.
Note: This scenario involves a more advanced statistical analysis known as ‘Logistic Regression’, not covered in either Descriptive or Inferential Statistics. The general premise of Logistic Regression is to determine whether a predictor variable has an effect on the outcome of a categorical response variable. In this scenario our categorical outcome is binary (no aggression, 0, versus aggression, 1). This makes this scenario more specifically a ‘Binomial Regression’, which is a subset of Logistic Regression. For a primer on how to conduct Binomial Regression in R, see:
https://bookdown.org/ndphillips/YaRrr/logistic-regression-with-glmfamily-binomial.html
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=81#h5p-17
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=81#h5p-18
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=81#h5p-19
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=81#h5p-20
To download, right-click and press “Save File As” or “Download Linked File”
Behavioural and Sensory Ecology Lab, Dr. James B. Barnett, Department of Zoology, Trinity College Dublin, Ireland
Chaloupka, S., Peignier, M., Stückler, S., Araya-Ajoy, Y., Walsh, P., Ringler, M., & Ringler, E. (2022). Repeatable territorial aggression in a Neotropical poison frog. Frontiers in ecology and evolution, 10, 881387.
Rodríguez, C., Fusani, L., Raboisson, G., Hödl, W., Ringler, E., & Canoine, V. (2022). Androgen responsiveness to simulated territorial intrusions in Allobates femoralis males: evidence supporting the challenge hypothesis in a territorial frog. General and comparative endocrinology, 326, 114046.
You are a visual ecologist studying warning colouration and mimicry in poison dart frogs. Your study system is the toxic Ameerega bilinguis and the non-toxic Allobates zaparo, a pair of sympatric terrestrial frog species native to the Ecuadorian Amazon. Ameerega bilinguis utilizes a multi-component warning signal, with a red dorsum, bright yellow limb-pit spots, and a bright blue belly. Allobates zaparo has evolved to mimic this colouration, but exhibits ‘imperfect mimicry’ – the exact quantitative properties of the color components are not perfectly matched. You collected 20 adult individuals of each species from the field and took color-calibrated photographs of each of their body regions. You then used the micaToolbox in ImageJ to simulate avian vision and compute the strength of the chromatic (i.e. hue) and achromatic (i.e. brightness) contrast of each of four color components (Front Spots, Back Spots, Dorsum, Venter) against a natural leaf-litter background, for each species. Visual contrast values are presented in the unit of ‘JND’, or ‘Just Noticeable Difference’, where higher values indicate that the color patch contrasts more strongly with the background. In other words, higher values mean that signal component is more conspicuous.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=83#h5p-21
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=83#h5p-22
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=83#h5p-23
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=83#h5p-24
To download, right-click and press “Save File As” or “Download Linked File”
Behavioural and Sensory Ecology Lab; Dr. James Barnett @ Trinity College Dublin
McEwen, B.L., Yeager, J.D., Kinley, I., Anderson, H.M., Barnett, J.B.B. (Under Review). Body posture and viewing angle modulate detectability and mimic fidelity in a poison frog system.
Darst, C. R., & Cummings, M. E. (2006). Predator learning favours mimicry of a less-toxic model in poison frogs. Nature, 440(7081), 208-211.
Stevens M, Párraga CA, Cuthill IC, Partridge JC, Troscianko TS. (2007). Using digital photography to study animal coloration. Biol. J. Linn. 90, 211-237
You are a herpetologist interested in biological invasions in urban habiats. The brown anole Anolis sagrei is invasive to Southern Florida, where it now competes with the native green anole Anolis carolinensis. Males of both species fight over territory in their now shared habitat. The males aggressively signal using a series of pushups to display their physical condition to other males. Males that perform more pushups are deemed more aggressive than males who perform fewer pushups You are wondering whether the invasive brown anoles are better space competitors than the native green anoles, and whether space competition in these species is linked to their aggressive behaviour. To test this, you transported wild-captured individuals of both species to the lab. You administered three rounds of an aggression assay, in which the lizard was presented with a mirror and recorded. Males perceive their reflection to be an intruder, and engage in pushup display behaviour. You recorded the number of pushups performed in each of three separate trials, then calculated an ‘average aggression’ level for each lizard. You then placed one native A. carolinensis and one invasive A. sagrei into an arena and recorded the area of space they occupied over the course of three days (cm3) in a variable called ‘space use’, as a measure of their ability to compete for space.
Analyze this dataset to see whether species ID or aggression level has an effect on space competition ability.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=85#h5p-25
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=85#h5p-26
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=85#h5p-27
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=85#h5p-28
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=85#h5p-29
To download, right-click and press “Save File As” or “Download Linked File”
Unpublished work – PhD Candidate Brendan McEwen
Farrell, W. J., & Wilczynski, W. (2006). Aggressive experience alters place preference in green anole lizards, Anolis carolinensis. Animal Behaviour, 71(5), 1155-1164.
Rodríguez, C., Fusani, L., Raboisson, G., Hödl, W., Ringler, E., & Canoine, V. (2022). Androgen responsiveness to simulated territorial intrusions in Allobates femoralis males: evidence supporting the challenge hypothesis in a territorial frog. General and comparative endocrinology, 326, 114046.
You are a sociobiologist investigating the genetic basis of sociability in fruit flies. A genetic screening study has identified several ‘candidate genes’ that may play a role in regulating social behaviour in Drosophila melanogaster. You decide to investigate the candidate gene Sec5, by performing a knockdown experiment. Male and female cohorts of flies were subjected to RNA Interference Gene Silencing (RNAi), effectively eliminating Sec5 gene activity in those individuals. Another genetically un-altered cohort of males and females were kept as controls. You assembled groups of same-sex flies into sociability arenas, and tracked their aggregation behaviour over time. A ‘sociability index’ score was calculated for each group, where higher scores indicate that the flies were more closely aggregated towards one another – a stronger signal of social grouping.
Analyze this dataset to see whether males and females differ in their levels of sociality, and whether the silencing of the Sec5 gene affected sociality in either males or females
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=87#h5p-30
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=87#h5p-31
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=87#h5p-32
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=87#h5p-33
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=87#h5p-34
To download, right-click and press “Save File As” or “Download Linked File”
Cognitive Ecology Lab, Dr. Reuven Dukas, McMaster University Department of Psychology, Neuroscience, & Behaviour https://psych.mcmaster.ca/dukas/index.htm
Torabi-Marashi, A. (2023). Investigating the genetic basis of natural variation in sociability within Drosophila melanogaster (Doctoral dissertation)
Scott, A. M., Dworkin, I., & Dukas, R. (2022). Evolution of sociability by artificial selection. Evolution, 76(3), 541-553
As a behavioural ecologist at the Hamilton Research Institute, you are delving into the interesting social dynamics of Neolamprologus pulcher, the group-living cichlid fish native to Lake Tanganyika in Africa. Your primary objective is to explore the impact of social rank on learning abilities within N. pulcher groups. In this captivating study, each social unit of N. pulcher comprises two dominant breeders and a varying number of subordinate helpers, often reaching up to 20 individuals. Your research unfolds through a series of three experiments, all documented in a comprehensive dataset. The dataset encompasses key variables, including fish ID, sex, size, and three critical aspects of learning:
Initial Learning (Experiment 1): Fish were individually trained to move a coloured disc (blue) to uncover a hidden food source. Data are in clomun “LearnedInitial”.
Associate Learning (Experiment 2): A second disc, coloured yellow and immovable, was introduced to evaluate the fish’s ability to associate it with food. Data are in column “LearnedAssos”.
Reverse Learning (Experiment 3): In this experiment, the colour of the disc that could be moved was switched, challenging the fish to adapt and reverse their learned behaviour. Data are in column “LearnedReverse”.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=89#h5p-35
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=89#h5p-36
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=89#h5p-37
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=89#h5p-38
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=89#h5p-39
As a researcher investigating the dietary habits of juvenile round goby (Neogobius melanostomus) in Hamilton Harbour, you’re delving into the intricacies of their feeding behavior during both day and night. Round goby, a small freshwater fish native to Eurasia, has become established in various regions across North America, including the Great Lakes. These goby, known for their voracious appetites and adaptability, play a significant role in local ecosystems as both predators and prey.
In Hamilton Harbour, juvenile round goby primarily prey on two main types of organisms: cladocerans and rotifers. Your dataset provides valuable insights into the stomach contents of these juveniles, revealing their consumption patterns across different times of the day. Recorded variables include the fish ID, the time of collection, and the quantities of cladocerans and rotifers found in each fish’s stomach.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=91#h5p-40
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=91#h5p-41
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=91#h5p-42
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=91#h5p-43
In this experiment, researchers investigated the dispersal behaviour of round goby in a controlled laboratory setting, exploring how it varies under day and night conditions. Additionally, the activity levels of the goby were recorded within the experimental tank. Each goby was categorized based on its size (Small, Medium, or Large) and whether it exhibited dispersal behaviour. The activity level of each goby was also measured. The dataset comprises crucial information on the size of each goby, their dispersal behaviour, and their activity levels within the experimental environment.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=93#h5p-44
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=93#h5p-45
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=93#h5p-46
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=93#h5p-47
Investigating the population dynamics of round goby (Neogobius melanostomus) in Lake Ontario, we delve into the biological characteristics of individuals sampled from this lake. The dataset includes details such as fish sex, length, and mass, crucial for understanding the demographics of this invasive species. By analyzing these attributes, we aim to gain insights into the distribution, growth patterns, and potential impacts of round goby within the lake’s ecosystem.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=95#h5p-48
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=95#h5p-49
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=95#h5p-50
In our study, we investigate the swimming behaviour of juvenile round goby (Neogobius melanostomus) using a swim tunnel experiment. This controlled environment allows us to observe two primary behaviours: active swimming, characterized by continuous movement and exploration, and station holding, where the fish maintain a relatively stationary position. By recording these behaviours, we aim to understand the activity patterns and preferences of round goby in response to varying conditions. Our dataset includes measurements of swimming and holding durations, as well as fish size categorization, providing insights into their locomotor dynamics and ecological interactions.
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=97#h5p-51
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=97#h5p-52
An interactive H5P element has been excluded from this version of the text. You can view it online here:
https://ecampusontario.pressbooks.pub/radpnb/?p=97#h5p-53
1
All data files referenced in this OER are available from the GitHub page here: https://github.com/HashemiScience/data4pnb/